Abstract
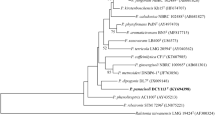
The novel species DCY115T was isolated from ginseng-cultivated soil in Gochang province, Republic of Korea. The isolated strain was assigned to the genus Paraburkholderia due to its 16S rRNA gene sequence proximity to Paraburkholderia xenovorans LB400T (98.8%), Paraburkholderia terricola LMG 20594T (98.4%), Paraburkholderia graminis C4D1MT (98.2%), Paraburkholderia rhynchosiae WSM3937T (98.1%), and Paraburkholderia phytofirmans PsJNT (98.1%). Strain DCY115T is gram-negative, facultative aerobic, rod-shaped, non-motile, non-flagellated, and oxidase and catalase positive. The predominant isoprenoid quinone of DCY115T is ubiquinone Q-8. The major cellular fatty acids are C16:0, cyclo-C17:0, cyclo-C19:0 ω8c, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c) and summed feature 8 (C18:1 ω7c and/or C18:1 ω6c). The major polar lipids include diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), and an unknown amino lipid (AL1). The genomic DNA G + C content is 61.3 mol%. Phenotypic tests and chemotaxonomic analysis place strain DCY115T in the genus Paraburkholderia. DNA–DNA hybridization values between strain DCY115T and closely related reference strains were lower than 51%. The low DNA relatedness data in combination with phylogenetic and biochemical tests showed that strain DCY115T could not be assigned to any recognized species. Finally, strain DCY115T showed antagonistic activity against Fusarium solani (KACC 44891T) and Cylindrocarpon destructans (KACC 44660T), which are two root rot fungal pathogens of ginseng. In conclusion, the results in this study support strain DCY115T as a novel species within the genus Paraburkholderia for which the name Paraburkholderia panacihumi is proposed. The type strain is DCY115T (= KCTC 52952T = JCM 32099T).


Similar content being viewed by others
References
Bauer A, Kirby W, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Choi KT (2008) Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng CA Meyer. Acta Pharmacol Sin 29:1109–1118
Choi GM, Im WT (2018) Paraburkholderia azotifigens sp. nov., a nitrogen-fixing bacterium isolated from paddy soil. Int J Syst Evol Microbiol 68:310–316
Dobritsa AP, Samadpour M (2016) Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol 66:2836–2846
Ezaki T. Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Evol Microbiol 39:224–229
Farh ME-A, Kim YJ, Van An H, Sukweenadhi J, Singh P, Huq MA, Yang DC (2015) Burkholderia ginsengiterrae sp. nov. and Burkholderia panaciterrae sp. nov., antagonistic bacteria against root rot pathogen Cylindrocarpon destructans, isolated from ginseng soil. Arch Microbiol 197:439–447
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416
Gao Z, Yuan Y, Xu L, Liu R, Chen M, Zhang C (2016) Paraburkholderia caffeinilytica sp. nov., isolated from the soil of a tea plantation. Int J Syst Evol Microbiol 66:4185–4190
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Kang JP, Nguyen NL, Kim YJ, Hoang VA, Bae KS, Yang DC (2015) Paralcaligenes ginsengisoli sp. nov., isolated from ginseng cultivated soil. Antonie Van Leeuwenhoek 108:619–626
Kim OS et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–176
Levine M, Epstein S, Vaughn R (1934) Differential reactions in the colon group of bacteria. Am J Public Health Nations Health 24:505–510
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Evol Microbiol 39:159–167
Mesbah NM, Whitman WB, Mesbah M (2011) 14-determination of the G + C content of prokaryotes. Methods Microbiol 38:299–324
Minnikin D, O’donnell A, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Pikovskaya R (1948) Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17:362–370
Rahman M, Punja ZK (2005) Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathology 95:1381–1390
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI Inc, Newark
Sawana A, Adeolu M, Gupta RS (2014) Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429
Schwyn B, Neilands J (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Yabuuchi E et al (1992) Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol 36:1251–1275
Acknowledgements
This study was supported by a Grant from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (KIPET No. 317007-3), Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Erko Stackebrandt.
The NCBI GenBank accession number for the 16S rRNA gene sequence of strain DCY115T is KY694400.
DPD (digital protologue database) Taxon Number TA00399.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huo, Y., Kang, JP., Kim, YJ. et al. Paraburkholderia panacihumi sp. nov., an isolate from ginseng-cultivated soil, is antagonistic against root rot fungal pathogen. Arch Microbiol 200, 1151–1158 (2018). https://doi.org/10.1007/s00203-018-1530-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1530-2