Abstract
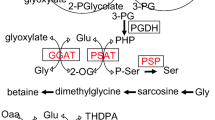
A link between carbon and nitrogen metabolism is important for serving as metabolic ancillary reactions. Here, we identified and characterized the alanine dehydrogenase gene in Aphanothece halophytica (ApalaDH) that is involved in alanine assimilation/dissimilation. Functional analysis revealed that ApalaDH encodes a bifunctional protein catalyzing the reversible reaction of pyruvate to l-alanine via its pyruvate reductive aminase (PvRA) activity, the reaction of l-alanine to pyruvate via its alanine oxidative dehydrogenase activity, and the non-reversible reaction of glyoxylate to glycine via its glyoxylate reductive aminase (GxRA) activity. Kinetic analysis showed the lowest affinity for pyruvate followed by l-alanine and glyoxylate with a Km of 0.22 ± 0.02, 0.72 ± 0.04, and 1.91 ± 0.43 mM, respectively. ApalaDH expression was upregulated by salt. Only PvRA and GxRA activities were detected in vivo and both activities increased about 1.2- and 2.7-fold upon salt stress. These features implicate that the assimilatory/dissimilatory roles of ApAlaDH are not only selective for l-alanine and pyruvate, but also, upon salt stress, can catabolize glyoxylate to generate glycine.






Similar content being viewed by others
Abbreviations
- ADH:
-
Alanine dehydrogenase
- PvRA:
-
Pyruvate reductive aminase activity
- ALD:
-
Alanine oxidative dehydrogenase activity
- GxRA:
-
Glyoxylate reductive aminase activity
- GDH:
-
Glycine oxidative dehydrogenase activity
References
Al Onazi M, Al-DAhain S, El-Ansary A, Marraiki N (2011) Isolation and characterization of Thielaviopsis paradoxa l-alanine dehydrogenase. Asian J Appl Sci. https://doi.org/10.3923/ajap.2011
Bellion E, Tan F (1987) An NAD+-dependent alanine dehydrogenase from a methylotrophic bacterium. Biochem J 244:565–570
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brunhuber NM, Blanchard JS (1994) The biochemistry and enzymology of amino acid dehydrogenases. Crit Rev Biochem Mol Biol 29:415–467
Caballero FJ, Cardenas J, Castillo F (1989) Purification and properties of l-alanine dehydrogenase of the phototrophic bacterium Rhodobacter capsulatus E1F1. J Bacteriol 171:3205–3210
Chowdhury EK, Saitoh T, Nagata S, Ashiuchi M, Misono H (1998) Alanine dehydrogenase from Enterobacter aerogenes: purification, characterization, and primary structure. Biosci Biotechnol Biochem 62:2357–2363
Giffin MM, Modesti L, Raab RW, Wayne LG, Sohaskey CD (2012) ald of Mycobacterium tuberculosis encodes both the alanine dehydrogenase and the putative glycine dehydrogenase. J Bacteriol 194:1045–1054
Hagins JM, Locy R, Silo-Suh L (2009) Isocitrate lyase supplies precursors for hydrogen cyanide production in a cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol 191:6335–6339
Hutter B, Singh M (1999) Properties of the 40 kDa antigen of Mycobacterium tuberculosis, a functional l-alanine dehydrogenase. Biochem J 343 Pt 3:669–672
June-Don B, Cho YJ, Kim DI, Lee DS, Shin HJ (2003) Purification and biochemical characterization of recombinant alanine dehydrogenase from Thermus caldophilus GK24. J Microbiol Biotechnol 13(4):628–631
Kuroda S, Tanizawa K, Sakamoto Y, Tanaka H, Soda K (1990) Alanine dehydrogenases from two Bacillus species with distinct thermostabilities: molecular cloning, DNA and protein sequence determination, and structural comparison with other NAD(P)(+)-dependent dehydrogenases. Biochemistry 29(4):1009–1015
Lahmi R, Sendersky E, Perelman A, Hagemann M, Forchhammer K, Schwarz R (2006) Alanine dehydrogenase activity is required for adequate progression of phycobilisome degradation during nitrogen starvation in Synechococcus elongatus PCC 7942. J Bacteriol 188:5258–5265
Müller P, Werner D (1982) Alanine dehydrogenase from bacteroids and free living cells of Rhizobium japonicum. Z Naturforsch C 37:927–936
Pernil R, Herrero A, Flores E (2010) Catabolic function of compartmentalized alanine dehydrogenase in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 192:5165–5172
Rowell P, Stewart WD (1975) Alanine dehydrogenase of the N2-fixing blue-green alga, Anabaena cylindrica. Arch Microbiol 107:115–124
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2 edn. Cold Spring Harbor Laboratory Press, Plainview
Sawa Y, Tani M, Murata K, Shibata H, Ochiai H (1994) Purification and characterization of alanine dehydrogenase from a cyanobacterium, Phormidium lapideum. J Biochem 116(5):995–1000
Schlupen C, Santos MA, Weber U, de Graaf A, Revuelta JL, Stahmann KP (2003) Disruption of the SHM2 gene, encoding one of two serine hydroxymethyltransferase isoenzymes, reduces the flux from glycine to serine in Ashbya gossypii. Biochem J 369:263–273
Schröder I, Vadas A, Johnson E, Lim S, Monbouquette HG (2004) A novel archaeal alanine dehydrogenase homologous to ornithine cyclodeaminase and mu-crystallin. J Bacteriol 186(22):7680–7689
Siranosian KJ, Ireton K, Grossman AD (1993) Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J Bacteriol 175:6789–6796
Smith MT, Emerich DW (1993) Alanine dehydrogenase from soybean nodule bacteroids: purification and properties. Arch Biochem Biophys 304(2):379–385
Vali Z, Kilar F, Lakatos S, Venyaminov SA, Zavodszky P (1980) l-alanine dehydrogenase from Thermus thermophilus. Biochim Biophys Acta 615:34–47
Waditee-Sirisattha R, Kageyama H, Sopun W, Tanaka Y, Takabe T (2014) Identification and upregulation of biosynthetic genes required for accumulation of Mycosporine-2-glycine under salt stress conditions in the halotolerant cyanobacterium Aphanothece halophytica. Appl Environ Microbiol 80(5):1763–1769. https://doi.org/10.1128/AEM.03729-13
Waditee-Sirisattha R, Kageyama H, Fukaya M, Rai V, Takabe T (2015) Nitrate and amino acid availability affects glycine betaine and mycosporine-2-glycine in response to changes of salinity in a halotolerant cyanobacterium Aphanothece halophytica. FEMS Microbiol Lett 362(23):fnv198. https://doi.org/10.1093/femsle/fnv198
Yoshida A, Freese E (1965) Enzymatic Properties of Alanine Dehydrogenase of Bacillus Subtilis. Biochim Biophys Acta 96:248–262
Acknowledgements
This study was funded by the research fund of the Research Institute of Meijo University and the Chemical Approaches for Food Applications Research Group, Faculty of Science, Chulalongkorn University.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure 1S. Effects of NaCl and KCl on ApAlaDH activity
. The following ApAlaDH enzymatic activities were assayed in the presence or absence of NaCl or KCl: A) and D) PvRA, B) and E) ALD, and C) and F) GxRA. The ApAlaDH activity measured in the absence of NaCl or KCl was set to 100%. The data is represented as the mean ± standard deviation from three independent experiments. (PPT 215 KB)
Rights and permissions
About this article
Cite this article
Phogosee, S., Hibino, T., Kageyama, H. et al. Bifunctional alanine dehydrogenase from the halotolerant cyanobacterium Aphanothece halophytica: characterization and molecular properties. Arch Microbiol 200, 719–727 (2018). https://doi.org/10.1007/s00203-018-1481-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1481-7