Abstract
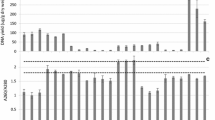
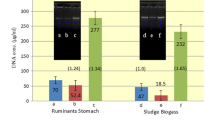
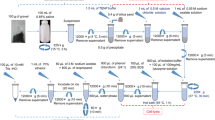
Various methods are available for DNA isolation from environmental samples. Because the chemical and biological composition of samples such as soil, sludge, or plant material is different, the effectiveness of DNA isolation can vary depending on the method applied and thus, have a substantial effect on the results of downstream analysis of the microbial community. Although the process of biogas formation is being intensely investigated, a systematic evaluation of kits for DNA isolation from material of biogas plants is still lacking. Since no DNA isolation kit specifically tailored for DNA isolation from sludge of biogas plants is available, this study compares five commercially available kits regarding their influence on downstream analyses such denaturing gradient gel electrophoresis (DGGE) and quantitative real-time PCR (qPCR). The results show that not all kits are equally suited for the DNA isolation from samples of different biogas plants, but highly reproducible DGGE fingerprints as well as qPCR results across the tested samples from biogas reactors using different substrate compositions could be produced using selected kits.




Similar content being viewed by others
References
An Y, Li G, Wu W, Huang J, He W, Zhu H (2014) Generation, collection and transportation, disposal and recycling of kitchen waste: a case study in Shanghai. Waste Manag Res 32:245–248. doi:10.1177/0734242X14521685
Bialek K, Kim J, Lee C, Collins G, Mahony T, O’Flaherty V (2011) Quantitative and qualitative analyses of methanogenic community development in high-rate anaerobic bioreactors. Water Res 45:1298–1308. doi:10.1016/j.watres.2010.10.010
Chen Y, Cheng JJ, Creamer KS (2008) Inhibition of anaerobic digestion process: a review. Bioresour Technol 99:4044–4064. doi:10.1016/j.biortech.2007.01.057
Cossu R, Masi S (2013) Re-thinking incentives and penalties: economic aspects of waste management in Italy. Waste Manag 33:2541–2547. doi:10.1016/j.wasman.2013.04.011
Fitzpatrick K, Kersh GJ, Massung RF (2010) Practical method for extraction of PCR-quality DNA from environmental soil samples. Appl Environ Microbiol 76:4571–4573. doi:10.1128/AEM.02825-09
Gagliano MC, Braguglia CM, Gallipoli A, Gianico A, Rossetti S (2014) Microbial diversity in innovative mesophilic/thermophilic temperature-phased anaerobic digestion of sludge. Environ Sci Pollut Res Int. doi:10.1007/s11356-014-3061-y
Grosskopf R, Janssen PH, Liesack W (1998) Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol 64:960–969
Holland PM, Abramson RD, Watson R, Gelfand DH (1991) Detection of specific polymerase chain reaction product by utilizing the 5′—3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 88:7276–7280
Hong SH, Bunge J, Leslin C, Jeon S, Epstein SS (2009) Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 3:1365–1373. doi:10.1038/ismej.2009.89
Inceoglu O, Hoogwout EF, Hill P, van Elsas JD (2010) Effect of DNA extraction method on the apparent microbial diversity of soil. Appl Environ Microbiol 76:3378–3382. doi:10.1128/AEM.02715-09
Ishii K, Fukui M (2001) Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. App Environ Microbiol 67:3753–3755. doi:10.1128/AEM.67.8.3753-3755.2001
Jang HM, Cho HU, Park SK, Ha JH, Park JM (2014a) Influence of thermophilic aerobic digestion as a sludge pre-treatment and solids retention time of mesophilic anaerobic digestion on the methane production, sludge digestion and microbial communities in a sequential digestion process. Water Res 48:1–14. doi:10.1016/j.watres.2013.06.041
Jang HM, Kim JH, Ha JH, Park JM (2014b) Bacterial and methanogenic archaeal communities during the single-stage anaerobic digestion of high-strength food wastewater. Bioresour Technol 165:174–182. doi:10.1016/j.biortech.2014.02.028
Khalid A, Arshad M, Anjum M, Mahmood T, Dawson L (2011) The anaerobic digestion of solid organic waste. Waste Manag 31:1737–1744. doi:10.1016/j.wasman.2011.03.021
Kim W, Lee S, Shin SG, Lee C, Hwang K, Hwang S (2012) Methanogenic community shift in anaerobic batch digesters treating swine wastewater. Water Res 44:4900–4907. doi:10.1016/j.watres.2010.07.029
Kim J, Kim W, Lee C (2013) Absolute dominance of hydrogenotrophic methanogens in full-scale anaerobic sewage sludge digesters. J Environ Sci (China) 25:2272–2280. doi:10.1016/s1001-0742(12)60299-x
Knauth S, Schmidt H, Tippkötter R (2013) Comparison of commercial kits for the extraction of DNA from paddy soils. Lett Appl Microbiol 56:222–228. doi:10.1111/lam.12038
LaMontagne MG, Michel FC Jr, Holden PA, Reddy CA (2002) Evaluation of extraction and purification methods for obtaining PCR-amplifiable DNA from compost for microbial community analysis. J Microbiol Methods 49:255–264. doi:10.1016/S0167-7012(01)00377-3
Li T, Mazéas L, Sghir A, Leblon G, Bouchez T (2009) Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol 11:889–904. doi:10.1111/j.1462-2920.2008.01810.x
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci 1125:171–189. doi:10.1196/annals.1419.019
Lv W, Zhang W, Yu Z (2013) Evaluation of system performances and microbial communities of two temperature-phased anaerobic digestion systems treating dairy manure. Bioresour Technol 143:431–438. doi:10.1016/j.biortech.2013.06.013
Mahmoudi N, Slater GF, Fulthorpe RR (2011) Comparison of commercial DNA extraction kits for isolation and purification of bacterial and eukaryotic DNA from PAH-contaminated soils. Can J Microbiol 57:623–628. doi:10.1139/w11-049
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Ovreås L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373
Schriewer A, Wehlmann A, Wuertz S (2011) Improving qPCR efficiency in environmental samples by selective removal of humic acids with DAX-8. J Microbiol Methods 85:16–21. doi:10.1016/j.mimet.2010.12.027
Sheffield VC, Cox DR, Lerman LS, Myers RM (1989) Attachment of a 40-base-pair G+ C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA 86(1):232–236
Stach JE, Bathe S, Clapp JP, Burns RG (2001) PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol Ecol 36:139–151
Stahl DA, Amann R (1991) Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematic. Wiley, Chichester, pp 205–248
Tebbe CC, Vahjen W (1993) Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol 59:2657–2665
Vissers EW, Bodelier PLE, Muyzer G, Laanbroek HJ (2009) A nested PCR approach for improved recovery of archaeal 16S rRNA gene fragments from freshwater samples. FEMS Microbiol Lett 298:193–198. doi:10.1111/j.1574-6968.2009.01718.x
von Wintzingerode F, Göbel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229. doi:10.1111/j.1574-6976.1997.tb00351.x
Williams J, Williams H, Dinsdale R, Guwy A, Esteves S (2013) Monitoring methanogenic population dynamics in a full-scale anaerobic digester to facilitate operational management. Bioresour Technol 140:234–242. doi:10.1016/j.biortech.2013.04.089
Yu Z, Morrison M (2004) Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812
Yu Y, Lee C, Kim J, Hwang S (2005) Analysis of community structures in anaerobic processes using a quantitative real-time PCR method. Water Sci Technol 52:85–91. doi:10.1002/bit.20347
Yu D, Kurola JM, Lähde K, Kymäläinen M, Sinkkonen A, Romantschuk M (2014) Biogas production and methanogenic archaeal community in mesophilic and thermophilic anaerobic co-digestion processes. J Environ Manage. doi:10.1016/j.jenvman.2014.04.025
Zhang X, Huang G (2014) Municipal solid waste management planning considering greenhouse gas emission trading under fuzzy environment. J Environ Manage 135:1–18. doi:10.1016/j.jenvman.2014.01.014
Acknowledgments
This study was supported by the Bundesministerium für Bildung und Forschung (BMBF) via the BioPara network (Grant 03SF0421C). We thank Bioreact GmbH (Troisdorf, Germany) and the Stadtentwässerung of Dresden, Germany for the provision of sludge samples and Tobias Kern (Technische Universität Dresden, Germany) for his assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Harald Huber.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Theiss, J., Rother, M. & Röske, K. Influence of DNA isolation method on the investigation of archaeal diversity and abundance in biogas plants. Arch Microbiol 198, 619–628 (2016). https://doi.org/10.1007/s00203-016-1221-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-016-1221-9