Abstract
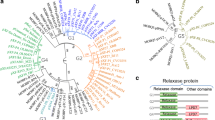
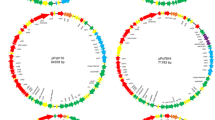
Recent genome analysis of Erwinia amylovora, the causal agent of fire blight disease on Rosaceae, has shown that the chromosome is highly conserved among strains and that plasmids are the principal source of genomic diversity. A new circular plasmid, pEA68, was found in E. amylovora strain 692 (LMG 28361), isolated in Poland from Sorbus (mountain ash) with fire blight symptoms. Annotation of the 68,763-bp IncFIIa-type plasmid revealed that it contains 79 predicted CDS, among which two operons (tra, pil) are associated with mobility. The plasmid is maintained stably in E. amylovora and does not possess genes associated with antibiotic resistance or known virulence genes. Curing E. amylovora strain 692 of pEA68 did not influence its virulence in apple shoots nor amylovoran synthesis. Of 488 strains of E. amylovora from seventeen countries, pEA68 was only found in two additional strains from Belgium. Although the spread of pEA68 is currently limited to Europe, pEA68 comprises, together with pEA72 and pEA78 both found in North America, a new plasmid family that spans two continents.





Similar content being viewed by others
References
Bellemann P, Bereswill S, Berger S, Geider K (1994) Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int J Biol Macromol 16(6):290–296
Bertani G (1951) Studies on lysogenesis. 1. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62(3):293–300
Bonn GW, van der Zwet T (2000) Distribution and economic importance of fire blight. In: Vanneste JL (ed) Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, pp 37–54
Bühlmann A, Dreo T, Rezzonico F, Pothier JF, Smits TH, Ravnikar M, Frey JE, Duffy B (2014) Phylogeography and population structure of the biologically invasive phytopathogen Erwinia amylovora inferred using minisatellites. Environ Microbiol 16:2112–2125
Carey AB, Pusey PL, Smith TJ, Loper JE, Stockwell VO (2011) Plasmid content of isolates of Erwinia amylovora from orchards in Washington and Oregon in the USA. Acta Hort 896:123–126
Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403
Falkenstein H, Zeller W, Geider K (1989) The 29 kb plasmid, common in strains of Erwinia amylovora, modulates development of fireblight symptoms. J Gen Microbiol 135:2643–2650
Foster GC, McGhee GC, Jones AL, Sundin GW (2004) Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl Environ Microbiol 70:7539–7544
Garcillán-Barcia MP, Francia MV, de la Cruz F (2009) The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687
Garcillán-Barcia MP, Alvarado A, de la Cruz F (2011) Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev 35:936–956
Gołębiewski M, Kern-Zdanowicz I, Zienkiewicz M, Adamczyk M, Żylińska J, Baraniak A, Gniadkowski M, Bardowski J, Cegłowski P (2007) Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum ß-lactamase gene bla (CTX-M-3). Antimicrob Agents Chemother 51:3789–3795
Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY (2011) Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119
Jock S, Donat V, López MM, Bazzi C, Geider K (2002) Following spread of fire blight in Western, Central and Southern Europe by molecular differentiation of Erwinia amylovora strains with PFGE analysis. Environ Microbiol 4:106–114
Jock S, Wensing A, Pulawska J, Drenova N, Dreo T, Geider K (2013) Molecular analyses of Erwinia amylovora strains isolated in Russia, Poland, Slovenia and Austria describing further spread of fire blight in Europe. Microbiol Res 168:447–454
Laurent J, Barny M, Katoujansky A, Dufriche P, Vanneste JL (1989) Characterization of a ubiquitous plasmid in Erwinia amylovora. Mol Plant Microbe Interact 2:160–164
Lelliot RA (1959) Fire blight of pears in England. Agriculture 56:564–568
Llop P, Bonaterra A, Peñalver J, López MM (2000) Development of a highly sensitive nested-PCR procedure using a single closed tube for detection of Erwinia amylovora in asymptomatic plant material. Appl Environ Microbiol 66:2071–2078
Llop P, Donat V, López MM, Cabrefiga J, Ruz L, Montesinos E, Palomo JL, Rodriguez M (2006) An Erwinia amylovora strain without plasmid pEA29 isolated in a nursery from hawthorn. Acta Hort 704:431–437
Llop P, Cabrefiga J, Smits THM, Dreo T, Barbe S, Pulawska J, Bultreys A, Blom J, Duffy B, Montesinos E, López MM (2011) Erwinia amylovora novel plasmid pEI70: complete sequence, biogeography, and role in aggressiveness in the fire blight phytopathogen. PLoS One 6:e28651
Llop P, Barbé S, López MM (2012) Functions and origin of plasmids in Erwinia species that are pathogenic to or epiphytically associated with pome fruit trees. Trees Struct Funct 26:31–46
Maes M, Orye K, Bobev S, Devreese B, Van Beeumen J, De Bruyn A, Busson R, Herdewijn P, Morreel K, Messens E (2001) Influence of amylovoran production on virulence of Erwinia amylovora and a different amylovoran structure in E. amylovora isolates from Rubus. Eur J Plant Pathol 107:839–844
Mann RA, Smits THM, Bühlmann A, Blom J, Goesmann A, Frey JE, Plummer KM, Beer SV, Luck J, Duffy B, Rodoni B (2013) Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core. PLoS One 8(2):e55644
McGhee GC, Jones AL (2000) Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Appl Environ Microbiol 66:4897–4907
McGhee GC, Foster GC, Jones AL (2002) Genetic diversity among Erwinia amylovora’s ubiquitous plasmid pEA29. Acta Hort 590:413–421
Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A (2003) GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31:2187–2195
Mohammadi M, Moltmann E, Zeller W, Geider K (2009) Characterisation of naturally occurring Erwinia amylovora strains lacking the common plasmid pEA29 and their detection with real-time PCR. Eur J Plant Pathol 124:293–302
Powney R, Smits THM, Sawbridge T, Frey B, Blom J, Frey JE, Plummer KM, Beer SV, Luck J, Duffy B, Rodoni B (2011) Genome sequence of an Erwinia amylovora strain with pathogenicity restricted to Rubus plants. J Bacteriol 193:785–786
Puławska J, Sobiczewski P (2012) Phenotypic and genetic diversity of Erwinia amylovora: the causal agent of fire blight. Trees Struct Funct 26:3–12
Puławska J, Kielak K, Sobiczewski P (2006) Phenotypic and genetic diversity of selected Erwinia amylovora strains from Poland. Acta Hort 704:439–444
Schnabel EL, Jones AL (1998) Instability of a pEA29 marker in Erwinia amylovora previously used for strain classification. Plant Dis 82:1334–1336
Sebaihia M, Bocsanczy AM, Biehl BS, Quail MA, Perna NT, Glasner JD, DeClerck GA, Cartinhour S, Schneider DJ, Bentley SD, Parkhill J, Beer SV (2010) Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J Bacteriol 192:2020–2021
Smits THM, Rezzonico F, Kamber T, Blom J, Goesmann A, Frey JE, Duffy B (2010) Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol Plant Microbe Interact 23:384–393
Smits THM, Guerrero-Prieto VM, Hernández-Escarcega G, Blom J, Goesmann A, Rezzonico F, Duffy B, Stockwell VO (2014) Whole-genome sequencing of Erwinia amylovora strains from Mexico detects SNPs in rpsL conferring streptomycin resistance and in the avrRpt2 effector altering host interactions. Genome Announc. http://genomea.asm.org/content/2/1/e01229-13.full.pdf+html
Thomson SV (2000) Epidemiology of fire blight. In: Vanneste JL (ed) Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, pp 9–36
Wang DP, Korban SS, Zhao YF (2009) The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora. Mol Plant Pathol 10:277–290
Zhou C, Yang YJ, Jong AY (1990) Mini-prep in 10 minutes. Biotechniques 8:172–173
Acknowledgments
We thank Jan van der Wolf (Plant Research International, Wageningen, The Netherlands), Svetoslav Bobev (Agricultural University, Plovdiv, Bulgaria), Said Sadallah (University of Skikda, Algeria), Anatoli Nikolaevich Evtushenkov (Belarusian State University, Minsk, Belarus), Alia Abed El-Baky Shoeib (Alexandria University, Egypt), Milda Vasinauskiene (Institute of Botany, Vilnius, Lithuania) and Maria López (IVIA, Valencia, Spain) for providing strains. Funding was provided by MARD (project HORhn8427/1/2013), Euphresco ERA-Net pilot project PhytFire (www.phytfire.org) and III 46008 (Ministry of Education, Science and Technological Development, Republic of Serbia).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ismail, E., Blom, J., Bultreys, A. et al. A novel plasmid pEA68 of Erwinia amylovora and the description of a new family of plasmids. Arch Microbiol 196, 891–899 (2014). https://doi.org/10.1007/s00203-014-1028-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-014-1028-5