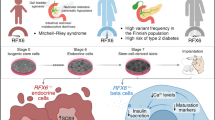
Abstract
Aims/hypothesis
Type 1 diabetes is a T cell-mediated autoimmune disease characterised by pancreatic beta cell destruction. In this study, we explored the pathogenic immune responses in initiation of type 1 diabetes and new immunological targets for type 1 diabetes prevention and treatment.
Methods
We obtained peripheral blood samples from four individuals with newly diagnosed latent autoimmune diabetes in adults (LADA) and from four healthy control participants. Single-cell RNA-sequencing (scRNA-seq) was performed on peripheral blood mononuclear cells to uncover transcriptomic profiles of early LADA. Validation was performed through flow cytometry in a cohort comprising 54 LADA, 17 adult-onset type 2 diabetes, and 26 healthy adults, matched using propensity score matching (PSM) based on age and sex. A similar PSM method matched 15 paediatric type 1 diabetes patients with 15 healthy children. Further flow cytometry analysis was performed in both peripheral blood and pancreatic tissues of non-obese diabetic (NOD) mice. Additionally, cell adoptive transfer and clearance assays were performed in NOD mice to explore the role of this monocyte subset in islet inflammation and onset of type 1 diabetes.
Results
The scRNA-seq data showed that upregulated genes in peripheral T cells and monocytes from early-onset LADA patients were primarily enriched in the IFN signalling pathway. A new cluster of classical monocytes (cluster 4) was identified, and the proportion of this cluster was significantly increased in individuals with LADA compared with healthy control individuals (11.93% vs 5.93%, p=0.017) and that exhibited a strong IFN signature marked by SIGLEC-1 (encoding sialoadhesin). These SIGLEC-1+ monocytes expressed high levels of genes encoding C-C chemokine receptors 1 or 2, as well as genes for chemoattractants for T cells and natural killer cells. They also showed relatively low levels of genes for co-stimulatory and HLA molecules. Flow cytometry analysis verified the elevated levels of SIGLEC-1+ monocytes in the peripheral blood of participants with LADA and paediatric type 1 diabetes compared with healthy control participants and those with type 2 diabetes. Interestingly, the proportion of SIGLEC-1+ monocytes positively correlated with disease activity and negatively with disease duration in the LADA patients. In NOD mice, the proportion of SIGLEC-1+ monocytes in the peripheral blood was highest at the age of 6 weeks (16.88%), while the peak occurred at 12 weeks in pancreatic tissues (23.65%). Adoptive transfer experiments revealed a significant acceleration in diabetes onset in the SIGLEC-1+ group compared with the SIGLEC-1− or saline control group.
Conclusions/interpretation
Our study identified a novel group of SIGLEC-1+ monocytes that may serve as an important indicator for early diagnosis, activity assessment and monitoring of therapeutic efficacy in type 1 diabetes, and may also be a novel target for preventing and treating type 1 diabetes.
Data availability
RNA-seq data have been deposited in the GSA human database (https://ngdc.cncb.ac.cn/gsa-human/) under accession number HRA003649.
Graphical Abstract








Similar content being viewed by others
Abbreviations
- CD11b:
-
αM-integrin
- cDC2:
-
Conventional type 2 dendritic cell
- DEG:
-
Differentially expressed gene
- FCM:
-
Flow cytometry
- GADA:
-
GAD65 antibody
- GDT:
-
γδ T cell
- GO:
-
Gene Ontology
- ICR:
-
Institute of Cancer Research
- LADA:
-
Latent autoimmune diabetes in adults
- Lin:
-
Lineage
- Ly6c:
-
Lymphocyte antigen 6 complex locus C
- MAIT:
-
Mucosal-associated invariant T cell
- MX1:
-
Myxovirus Resistance 1
- NK:
-
Natural killer cell
- NKT:
-
Natural killer T cell
- NOD:
-
Non-obese diabetic
- PBMC:
-
Peripheral blood mononuclear cell
- pDC:
-
Plasmacytoid dendritic cell
- PLXNB2:
-
Plexin B2
- PSM:
-
Propensity score matching
- scRNA-seq:
-
Single-cell RNA-sequencing
- Sema4D:
-
Semaphorin 4D
- SIGLEC-1:
-
Sialoadhesin
- STAT1:
-
Signal transducer and activator of transcription 1
- Teff:
-
Effector T cell
- Tem:
-
Effector memory T cell
- Treg:
-
Regulatory T cell
- UMAP:
-
Uniform Manifold Approximation and Projection
References
Green A, Hede SM, Patterson CC et al (2021) Type 1 diabetes in 2017: global estimates of incident and prevalent cases in children and adults. Diabetologia 64(12):2741–2750. https://doi.org/10.1007/s00125-021-05571-8
Vanderniet JA, Jenkins AJ, Donaghue KC (2022) Epidemiology of type 1 diabetes. Curr Cardiol Rep 24(10):1455–1465. https://doi.org/10.1007/s11886-022-01762-w
American Diabetes Association Professional Practice Committee (2022) 2. Classification and diagnosis of diabetes: standards of medical care in diabetes – 2022. Diabetes Care 45(Suppl 1):S17–S38. https://doi.org/10.2337/dc22-S002
Hu J, Zhang R, Zou H, Xie L, Zhou Z, Xiao Y (2022) Latent autoimmune diabetes in adults (LADA): from immunopathogenesis to immunotherapy. Front Endocrinol (Lausanne) 13:917169. https://doi.org/10.3389/fendo.2022.917169
Bradshaw EM, Raddassi K, Elyaman W et al (2009) Monocytes from patients with type 1 diabetes spontaneously secrete proinflammatory cytokines inducing Th17 cells. J Immunol 183(7):4432–4439. https://doi.org/10.4049/jimmunol.0900576
Foss-Freitas MC, Foss NT, Donadi EA, Foss MC (2006) In vitro TNF-α and IL-6 production by adherent peripheral blood mononuclear cells obtained from type 1 and type 2 diabetic patients evaluated according to the metabolic control. Ann NY Acad Sci 1079:177–180. https://doi.org/10.1196/annals.1375.027
Ren X, Mou W, Su C et al (2017) Increase in peripheral blood intermediate monocytes is associated with the development of recent-onset type 1 diabetes mellitus in children. Int J Biol Sci 13(2):209–218. https://doi.org/10.7150/ijbs.15659
Diana J, Simoni Y, Furio L et al (2013) Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat Med 19(1):65–73. https://doi.org/10.1038/nm.3042
Carrero JA, Calderon B, Towfic F, Artyomov MN, Unanue ER (2013) Defining the transcriptional and cellular landscape of type 1 diabetes in the NOD mouse. PLoS One 8(3):e59701. https://doi.org/10.1371/journal.pone.0059701
Li Q, Xu B, Michie SA, Rubins KH, Schreriber RD, McDevitt HO (2008) Interferon-α initiates type 1 diabetes in nonobese diabetic mice. Proc Natl Acad Sci USA 105(34):12439–12444. https://doi.org/10.1073/pnas.0806439105
Ferreira RC, Guo H, Coulson RM et al (2014) A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes 63(7):2538–2550. https://doi.org/10.2337/db13-1777
Kallionpaa H, Elo LL, Laajala E et al (2014) Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes 63(7):2402–2414. https://doi.org/10.2337/db13-1775
Lundberg M, Krogvold L, Kuric E, Dahl-Jorgensen K, Skog O (2016) Expression of interferon-stimulated genes in insulitic pancreatic islets of patients recently diagnosed with type 1 diabetes. Diabetes 65(10):3104–3110. https://doi.org/10.2337/db16-0616
Fabris P, Betterle C, Greggio NA et al (1998) Insulin-dependent diabetes mellitus during alpha-interferon therapy for chronic viral hepatitis. J Hepatol 28(3):514–517. https://doi.org/10.1016/s0168-8278(98)80328-0
Crow MK (2010) Type I interferon in organ-targeted autoimmune and inflammatory diseases. Arthritis Res Ther 12(Suppl 1):S5. https://doi.org/10.1186/ar2886
Nakamura K, Kawasaki E, Imagawa A et al (2011) Type 1 diabetes and interferon therapy: a nationwide survey in Japan. Diabetes Care 34(9):2084–2089. https://doi.org/10.2337/dc10-2274
Meyer S, Woodward M, Hertel C et al (2016) AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell 166(3):582–595. https://doi.org/10.1016/j.cell.2016.06.024
Zhou Z, Xiang Y, Ji L et al (2013) Frequency, immunogenetics, and clinical characteristics of latent autoimmune diabetes in China (LADA China study): a nationwide, multicenter, clinic-based cross-sectional study. Diabetes 62(2):543–550. https://doi.org/10.2337/db12-0207
Alberti KG, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 15(7):539–553. https://doi.org/10.1002/(SICI)1096-9136(199807)15:7%3c539::AID-DIA668%3e3.0.CO;2-S
World Health Organization (2011) Use of glycated haemoglobin (HbA1c) in the diagnosis of diabetes mellitus: abbreviated report of a WHO consultation. World Health Organization, Geneva
Dura B, Choi JY, Zhang K et al (2019) scFTD-seq: freeze-thaw lysis based, portable approach toward highly distributed single-cell 3′ mRNA profiling. Nucleic Acids Res 47(3):e16. https://doi.org/10.1093/nar/gky1173
Satija R, Farrell JA, Gennert D, Schier AF, Regev A (2015) Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33(5):495–502. https://doi.org/10.1038/nbt.3192
Stuart T, Butler A, Hoffman P et al (2019) Comprehensive integration of single-cell data. Cell 177(7):1888-1902 e1821. https://doi.org/10.1016/j.cell.2019.05.031
Perez RK, Gordon MG, Subramaniam M et al (2022) Single-cell RNA-seq reveals cell type-specific molecular and genetic associations to lupus. Science 376(6589):eabf1970. https://doi.org/10.1126/science.abf1970
Yazar S, Alquicira-Hernandez J, Wing K et al (2022) Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376(6589):eabf3041. https://doi.org/10.1126/science.abf3041
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Andreatta M, Carmona SJ (2021) UCell: robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J 19:3796–3798. https://doi.org/10.1016/j.csbj.2021.06.043
Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R (2020) Cell PhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc 15(4):1484–1506. https://doi.org/10.1038/s41596-020-0292-x
Catrina AM, Popa MA, Vacaru AM, Fenyo IM (2021) Inflammatory status of the pancreas in NOD mice that do not develop overt diabetes. Rom J Morphol Embryol 62(1):109–115. https://doi.org/10.47162/RJME.62.1.10
Domingues A, Sartori A, Golim MA et al (2011) Prevention of experimental diabetes by Uncaria tomentosa extract: Th2 polarization, regulatory T cell preservation or both? J Ethnopharmacol 137(1):635–642. https://doi.org/10.1016/j.jep.2011.06.021
Rostami MR, Bradic M (2021) The derepression of transposable elements in lung cells is associated with the inflammatory response and gene activation in idiopathic pulmonary fibrosis. Mob DNA 12(1):14. https://doi.org/10.1186/s13100-021-00241-3
Lepelley A, Della Mina E, Van Nieuwenhove E et al (2021) Enhanced cGAS-STING-dependent interferon signaling associated with mutations in ATAD3A. J Exp Med 218(10):e20201560. https://doi.org/10.1084/jem.20201560
Kuo PT, Zeng Z, Salim N, Mattarollo S, Wells JW, Leggatt GR (2018) The role of CXCR3 and its chemokine ligands in skin disease and cancer. Front Med (Lausanne) 5:271. https://doi.org/10.3389/fmed.2018.00271
Zhang Y, Lazarus J, Steele NG et al (2020) Regulatory T-cell depletion alters the tumor microenvironment and accelerates pancreatic carcinogenesis. Cancer Discov 10(3):422–439. https://doi.org/10.1158/2159-8290.Cd-19-0958
Bourgoin P, Biechele G, Ait Belkacem I, Morange PE, Malergue F (2020) Role of the interferons in CD64 and CD169 expressions in whole blood: relevance in the balance between viral- or bacterial-oriented immune responses. Immun Inflamm Dis 8(1):106–123. https://doi.org/10.1002/iid3.289
Pino M, Erkizia I, Benet S et al (2015) HIV-1 immune activation induces Siglec-1 expression and enhances viral trans-infection in blood and tissue myeloid cells. Retrovirology 12:37. https://doi.org/10.1186/s12977-015-0160-x
Yang D, Tong L, Wang D, Wang Y, Wang X, Bai C (2010) Roles of CC chemokine receptors (CCRs) on lipopolysaccharide-induced acute lung injury. Respir Physiol Neurobiol 170(3):253–259. https://doi.org/10.1016/j.resp.2010.02.002
Soday L, Potts M, Hunter LM et al (2021) Comparative cell surface proteomic analysis of the primary human T cell and monocyte responses to type I interferon. Front Immunol 12:600056. https://doi.org/10.3389/fimmu.2021.600056
Affandi AJ, Olesek K, Grabowska J et al (2021) CD169 defines activated CD14+ monocytes with enhanced CD8+ T cell activation capacity. Front Immunol 12:697840. https://doi.org/10.3389/fimmu.2021.697840
Junqueira C, Crespo A, Ranjbar S et al (2022) FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature 606(7914):576–584. https://doi.org/10.1038/s41586-022-04702-4
York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, Lafyatis R (2007) A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum 56(3):1010–1020. https://doi.org/10.1002/art.22382
Biesen R, Demir C, Barkhudarova F et al (2008) Sialic acid-binding Ig-like lectin 1 expression in inflammatory and resident monocytes is a potential biomarker for monitoring disease activity and success of therapy in systemic lupus erythematosus. Arthritis Rheum 58(4):1136–1145. https://doi.org/10.1002/art.23404
Xie J, Wang Z, Wang W (2020) Semaphorin 4D induces an imbalance of Th17/Treg cells by activating the aryl hydrocarbon receptor in ankylosing spondylitis. Front Immunol 11:2151. https://doi.org/10.3389/fimmu.2020.02151
Atkin-Smith GK, Miles MA, Tixeira R et al (2019) Plexin B2 is a regulator of monocyte apoptotic cell disassembly. Cell Rep 29(7):1821-1831e3. https://doi.org/10.1016/j.celrep.2019.10.014
Tan T, Xiang Y, Deng C et al (2022) Variable frequencies of peripheral T-lymphocyte subsets in the diabetes spectrum from type 1 diabetes through latent autoimmune diabetes in adults (LADA) to type 2 diabetes. Front Immunol 13:974864. https://doi.org/10.3389/fimmu.2022.974864
Tassiulas I, Hu X, Ho H et al (2004) Amplification of IFN-α-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat Immunol 5(11):1181–1189. https://doi.org/10.1038/ni1126
Chaimowitz NS, Ebenezer SJ, Hanson IC, Anderson M, Forbes LR (2020) STAT1 gain of function, type 1 diabetes, and reversal with JAK inhibition. N Engl J Med 383(15):1494–1496. https://doi.org/10.1056/NEJMc2022226
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Acknowledgements
We would like to thank X. Jiang of the Third Military Medical University, China, for his help in the experiments. We also thank all the participants who made this research possible.
Data availability
RNA-seq data have been deposited in the GSA human database (https://ngdc.cncb.ac.cn/gsa-human/) under accession number HRA003649.
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers 81570694 and 82000417) and the Natural Science Foundation of Shandong (grant number ZR2014HL029).
Authors’ relationships and activities
MQG, FW (Fei Wang), YJD, FW (Fang Wang), LLX, YC, RL, YGW and SFL are current employees at the Affiliated Hospital of Qingdao University, China. The other authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
SFL, YGW and JZ were responsible for study conception and design. JNC, CW, YJD, FW (Fei Wang), FW (Fang Wang), LZ, HG, JJZ, RL and SKL were responsible for data acquisition and analysis. MQG, LLX and YC performed the data interpretation. MQG, LLX, YC and FW (Fei Wang) drafted the article. SFL, YGW, JZ, JNC, CW, YJD, FW (Fang Wang), LZ, HG, JJZ, RL and SKL critically reviewed the article. All authors approved the final version of this manuscript to be published. SFL is responsible for the integrity of the work as a whole.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, M., Guo, H., Zhu, J. et al. A novel subpopulation of monocytes with a strong interferon signature indicated by SIGLEC-1 is present in patients with in recent-onset type 1 diabetes. Diabetologia 67, 623–640 (2024). https://doi.org/10.1007/s00125-024-06098-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-024-06098-4