Abstract
Aims/hypothesis
Mitophagy, the selective autophagy of mitochondria, is essential for maintenance of mitochondrial function. Recent studies suggested that defective mitophagy in beta cells caused diabetes. However, because of technical difficulties, the development of a convenient and reliable method to evaluate mitophagy in beta cells in vivo is needed. The aim of this study was to establish beta cell-specific mitophagy reporter mice and elucidate the role of mitophagy in beta cell function under metabolically stressed conditions induced by a high-fat diet (HFD).
Methods
Mitophagy was assessed using newly generated conditional mitochondrial matrix targeting mitophagy reporter (CMMR) mice, in which mitophagy can be visualised specifically in beta cells in vivo using a fluorescent probe sensitive to lysosomal pH and degradation. Metabolic stress was induced in mice by exposure to the HFD for 20 weeks. The accumulation of dysfunctional mitochondria was examined by staining for functional/total mitochondria and reactive oxygen species (ROS) using specific fluorescent dyes and antibodies. To investigate the molecular mechanism underlying mitophagy in beta cells, overexpression and knockdown experiments were performed. HFD-fed mice were examined to determine whether chronic insulin treatment for 6 weeks could ameliorate mitophagy, mitochondrial function and impaired insulin secretion.
Results
Exposure to the HFD increased the number of enlarged (HFD-G) islets with markedly elevated mitophagy. Mechanistically, HFD feeding induced severe hypoxia in HFD-G islets, which upregulated mitophagy through the hypoxia-inducible factor 1-ɑ (Hif-1ɑ)/BCL2 interacting protein 3 (BNIP3) axis in beta cells. However, HFD-G islets unexpectedly showed the accumulation of dysfunctional mitochondria due to excessive ROS production, suggesting an insufficient capacity of mitophagy for the degradation of dysfunctional mitochondria. Chronic administration of insulin ameliorated hypoxia and reduced ROS production and dysfunctional mitochondria, leading to decreased mitophagy and restored insulin secretion.
Conclusions/interpretation
We demonstrated that CMMR mice enabled the evaluation of mitophagy in beta cells. Our results suggested that metabolic stress induced by the HFD caused the aberrant accumulation of dysfunctional mitochondria, which overwhelmed the mitophagic capacity and was associated with defective maintenance of mitochondrial function and impaired insulin secretion.
Graphical abstract

Similar content being viewed by others

Introduction
Type 2 diabetes occurs when pancreatic beta cells fail to meet the insulin demand from target tissues [1]. Although the amount of serum insulin depends on the secretory function of pancreatic beta cells and beta cell mass, cumulative evidence suggests that impaired glucose-stimulated insulin secretion (GSIS) triggers the onset of type 2 diabetes [2,3,4]. Insulin secretion from pancreatic beta cells depends on mitochondria that convert signals of elevated blood glucose into the increase in ATP, which triggers the exocytosis of insulin-containing granules [5, 6]. Increased blood glucose levels facilitate the glycolytic pathway to synthesise ATP and supply energy sources for mitochondrial metabolism. In mitochondria, metabolites of the glycolytic pathway stimulate the tricarboxylic acid (TCA) cycle, which drives ATP synthesis through oxidative phosphorylation. During oxidative phosphorylation, reactive oxygen species (ROS) are generated as an unavoidable byproduct, indicating that mitochondria are constitutively exposed to oxidative stress [3]. Because damaged mitochondria are detrimental to cellular homeostasis, their elimination is essential to prevent beta cell dysfunction. The damaged components of mitochondria are segregated as dysfunctional mitochondria and eliminated through autophagy. During mitochondrial autophagy (hereafter referred to as mitophagy), a membrane sac termed the isolation membrane is generated and engulfs dysfunctional mitochondria to form autophagosomes. Subsequently, the autophagosomes fuse with lysosomes, and dysfunctional mitochondria are degraded by lysosomal enzymes [7, 8]. In beta cells, the disruption of autophagic degradation by genetic ablation of autophagy related 7 (Atg7; an indispensable protein for autophagy) resulted in the accumulation of abnormal mitochondria, impaired GSIS and hyperglycaemia [9, 10]. In addition, corresponding phenotypes were reported in mice lacking proteins required for autophagy/mitophagy regulation [11,12,13], indicating that maintenance of mitochondrial function through mitophagy is important to preserve the beta cell function and euglycaemia.
Although previous mitophagy studies in beta cells used biochemical, immunohistochemical and electron microscopic techniques, pH-sensitive fluorescent probes have been applied to detect mitophagy in other cell types [14,15,16,17]. Because dysfunctional mitochondria inevitably experience changes in their pH environment when being delivered from the cytosol to lysosomes during mitophagy, the expression of pH-sensitive probes in mitochondria enables the detection of mitochondria degradation in lysosomes. The expression of tandemly ligated mCherry–enhanced GFP (EGFP) in mitochondria is a convenient method to detect mitophagy. Under neutral conditions, EGFP and mCherry fluoresce green and red, respectively. Moreover, EGFP is rapidly quenched and degraded, but mCherry fluorescence remains stable in lysosomes. As a result, mitochondria degraded in lysosomes show EGFP-negative but mCherry-positive signals [18], enabling the investigation of mitophagy in vivo [15, 19]. However, no report has evaluated mitophagy in beta cells using these probes in vivo. Thus, we developed a new fluorescent probe to elucidate the role of mitophagy in beta cell function under the metabolically stressed condition.
Here, we generated a beta cell-specific mitophagy reporter mouse using our recently developed mitophagy reporter (conditional mitochondrial matrix targeting mitophagy reporter [CMMR]) probe. CMMR mice revealed that high-fat diet (HFD) exposure caused the aberrant accumulation of dysfunctional mitochondria despite the upregulation of mitophagy through the hypoxia/hypoxia-inducible factor 1-ɑ (Hif-1ɑ)/BCL2 interacting protein 3 (BNIP3) axis in beta cells. Our results suggested that the generation of dysfunctional mitochondria overwhelmed the mitophagy capacity in beta cells in HFD-fed mice, leading to decreased mitochondrial function and impaired insulin secretion.
Methods
Animals
Cmmrflox/+ mice [17] and RIP-Cre mice [20] were used to generate CMMR mice. Pink1 knockout mice (with deletion of the gene encoding PTEN-induced kinase 1 [PINK1]) were obtained from the Jackson Laboratory (no. 017946). All mice were maintained in the C57BL/6 background purchased from Sankyo Labo Service (Japan), housed under a 12 h light/12 h dark cycle in climate-controlled facilities and fed either standard (CE-2; Clea Japan, Japan) or 60% fat rodent food (D12492; Research Diets, USA). Male mice were used for all experiments in this study. HFD feeding started at 6 weeks old. Mice were fasted for 16 h before being subjected to oral glucose tolerance tests with 1.25 g glucose/kg body weight, as described previously [13]. For chronic insulin treatments, 0.36 U/day Humulin R (Eli Lilly, USA) or volume-matched saline (154 mmol/l NaCl) was infused continuously using an osmotic minipump (ALZET, USA). The infusion was started after 14 weeks of HFD exposure, and mice were fed the HFD for an additional 6 weeks. For the hypoxia assay in isolated islets, pimonidazole (60 mg/kg body weight, Hypoxyprobe, USA) was injected 2 h before preparation [21]. Pancreatic islets of Langerhans cells were prepared as described previously [13, 22]. Animal experiments were approved by the Kyorin University Animal Care Committee (permission no. 18, 238).
Flux assay
Isolated islets were placed onto XF96 islet capture plates at 30 islets/well for control and size-matched islets from HFD-fed mice (HFD-SM) or ten islets/well for enlarged islets from HFD-fed mice (HFD-G). Following a 1 h incubation with XF RPMI medium (Agilent Technologies, USA) containing 2 mmol/l glucose and 0.1% FCS at 37°C, the plates were transferred to an XF96 analyser (Seahorse Bioscience, USA). Oxygen consumption rates (OCRs) were measured at 37°C in the absence and presence of 20 mmol/l glucose, 3.5 μmol/l oligomycin and a mixture of 1.5 μmol/l rotenone and antimycin A. The results were normalised to the baseline OCR.
Imaging analysis
Pancreatic sections were prepared as described previously [23]. To identify mitophagy signals, the mCherry signal intensity was subtracted from the EGFP signal in each pixel, and the subtracted images were converted to binary images by thresholding. Dilate and erode filters were applied to remove minimal noise. The ‘Analyze particle’ function in ImageJ v.1.53 (https://imagej.nih.gov/ij/) was used to extract and identify mitophagy signals. The total mitochondrial area was determined using images generated from mCherry and EGFP images with the ‘Max’ function in ImageJ. The numbers of mitophagy signals were normalised to the total mitochondrial area.
ROS measurements using MitoSOX, functional mitochondria imaging using MitoTracker Red CM-H2Xros (MTR) and immunostaining were performed as described previously [13, 22].
For electron microscopy analysis, beta cells cultured on slide glass were treated with 1 mmol/l deferiprone (DFP) for 12 h, followed by an additional 12 h treatment with 30 nmol/l bafilomycin A1 and 1 mmol/l DFP as described previously [24]. Isolated islets were cultured with 30 nmol/l bafilomycin A1 for 24 h. Cells and islets were analysed as described previously [13]. The total numbers of mitochondria and of those located in autophagosomes were manually counted.
Plasmids and recombinant adenoviruses
Plasmids and recombinant adenoviruses were generated using standard procedures. See the electronic supplementary material (ESM) Methods for further details.
Quantitative RT-PCR
Total RNA was extracted from isolated islets and quantitative RT-PCR was performed using a LightCycler 480 (Roche, Switzerland). See the ESM Methods for further details.
Insulin secretion assay and immunoblotting
Insulin secretion assays and immunoblotting were performed as described previously [22]. Antibodies used in this study are listed in ESM Table 1. OxyBlot assays were performed in accordance with the manufacturer’s instructions (Merck, Germany).
Islet size measurements
All islets isolated from control or HFD-fed mice were imaged under stereomicroscopy. See the ESM Methods for further details.
Min6 cells
Min6 cells, a gift from J. Miyazaki (Osaka University, Osaka, Japan) at passages 20 to 30, were cultured as described previously [22].
Statistical analysis
Data are expressed as the mean±SEM. Statistical analysis was performed using scipy.stats v.1.8.0 (https://scipy.org) and statmodels.stats v.0.13.0 (https://www.statsmodels.org) libraries. The equality of variance and normality were tested in all experiments. The significance of differences between datasets was assessed using statistical tests as indicated. A p value less than 0.05 was considered statistically significant.
Results
Mitophagy detection using a CMMR probe in pancreatic beta cells
To evaluate mitophagy in pancreatic beta cells, we used our recently developed fluorescent probe (CMMR probe) [17]. The CMMR probe was designed to express mCherry–EGFP in the mitochondrial matrix using a tandem mitochondria-targeting sequence derived from cytochrome c oxidase IV (Fig. 1a). We generated CMMR mice (Cmmrflox/+;Cre) by crossing Cmmrflox/+ mice with RIP-Cre mice expressing Cre recombinase in pancreatic beta cells [20]. CMMR mice appeared grossly normal and showed normal glucose tolerance against orally administered glucose. The total insulin content and GSIS in islets isolated from CMMR mice were equivalent to those in control islets (ESM Fig. 1), suggesting that the expression of CMMR probes did not disrupt mitochondrial function in beta cells.
Characterisation of the CMMR probe to evaluate mitophagy in pancreatic beta cells. (a) Scheme of the mitophagy-monitoring system. Mitochondria in the cytosol show both EGFP and mCherry fluorescence, whereas mitochondria delivered to lysosomes during mitophagy emit mCherry signals only. (b) A representative image of pancreatic sections from CMMR mice (green, EGFP; red, mCherry) immunostained for insulin (blue). (c) A representative image of CMMR beta cells (green, EGFP; red, mCherry) immunostained for the mitochondrial marker Tom20 (blue). (d) Representative electron micrographs of cultured beta cells treated with 30 nmol/l bafilomycin A1 alone (Control) or 30 nmol/l bafilomycin A1 and 1 mmol/l DFP (DFP). Arrowheads indicate mitochondria located in autophagosomes. (e) The number of mitochondria detected by electron microscopy as in (d) was quantified as the ratio of the number of mitochondria located in autophagosomes to the total number of mitochondria in each image (n=63 and 58 for control and DFP, respectively). ***p<0.001, Mann–Whitney U test. (f) CMMR beta cells were treated with 1 mmol/l DFP for 18 h in the presence or absence of 50 μmol/l CQ. (g) The relative numbers of EGFP(−)/mCherry(+) mitophagy signals in (f) were quantified (n=24, 19 and 15 images for control, DFP and DFP + CQ, respectively). ***p<0.001, Mann–Whitney U test, p values were adjusted by Holm’s method. (h) CMMR beta cells (green, EGFP; red, mCherry) starved for 3 h were immunostained for the lysosome marker LAMP1 (blue). (i) Representative images of CMMR beta cells (green, EGFP; red, mCherry) expressing control shRNA and TagRFP657 (blue) or Atg7 shRNA and TagRFP657. TagRFP657 was used as an infection marker. (j) The relative numbers of EGFP(−)/mCherry(+) mitophagy signals in CMMR beta cells expressing TagRFP657 together with control or Atg7 shRNA were quantified (n=16 and 24 images for control and Atg7, respectively). ***p<0.001, Student’s unpaired t test. Bars, 50 μm (b), 5 μm (c, f, h, i) and 2 μm (d)
We first characterised the CMMR probe in beta cells to examine whether it is a reliable marker for mitophagy. Frozen sections were prepared from pancreatic tissues isolated from CMMR mice and immunostained for insulin. As shown in Fig. 1b, mCherry and EGFP signals derived from the CMMR probe were detected in beta cells labelled with the anti-insulin antibody (89.5±1.0% of beta cells expressed CMMR probe, n=4098 beta cells in 16 islets from three mice). The localisation of the CMMR probe in beta cells was studied by immunostaining of beta cells cultured from CMMR mice (CMMR beta cells). As shown in Fig. 1c and ESM Fig. 2, signals derived from the CMMR probe were colocalised with the mitochondrial marker protein, translocase of outer mitochondrial membrane 20 (Tom20). Next, we aimed to detect mitophagy using the CMMR probe and electron microscopy, a classical and reliable method for mitophagy detection. Cultured beta cells were treated with the mitophagy inducer DFP [18] in the presence of bafilomycin A1 to inhibit the fusion of mitochondria-containing autophagosomes with lysosomes. Electron microscopy analysis revealed that DFP treatment significantly increased the number of mitochondria located in autophagosomes (Fig. 1d,e). Moreover, DFP treatment significantly increased the number of EGFP-negative but mCherry-positive [EGFP(−)/mCherry(+)] punctate signals in CMMR beta cells (Fig. 1f,g). To examine whether EGFP(−)/mCherry(+) signals were derived from degrading mitochondria in lysosomes, we next performed the immunostaining of CMMR beta cells for the lysosomal marker protein lysosomal-associated membrane protein 1 (LAMP1). As shown in Fig. 1h, EGFP(−)/mCherry(+) but not EGFP(+)/mCherry(+) signals were colocalised with LAMP1. In addition, the neutralisation of lysosomal pH using chloroquine (CQ) suppressed DFP-induced EGFP(−)/mCherry(+) signals (Fig. 1f,g). To examine whether EGFP(−)/mCherry(+) signals were mediated by autophagy, we used short hairpin RNA (shRNA) against Atg7. The Atg7 shRNA effectively reduced Atg7 expression in beta cell-derived clonal Min6 cells and cultured beta cells (ESM Fig. 3) and significantly suppressed the number of EGFP(−)/mCherry(+) signals in CMMR beta cells (Fig. 1i,j). These results indicated that EGFP(−)/mCherry(+) signals derived from the CMMR probe were produced by mitochondria being degraded during mitophagy (hereafter referred to as mitophagy signals).
HFD feeding upregulated mitophagy, especially in HFD-G islets
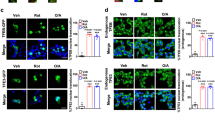
Increased autophagic flux was reported in obese mice [25]. In contrast, defective mitophagy was implicated in the pathogenesis of diabetes [11,12,13]. These results prompted us to hypothesise that diabetes affects mitophagy in beta cells. To test this hypothesis, CMMR mice were fed the HFD for 20 weeks, and CMMR signals were examined. HFD feeding induced glucose intolerance accompanied by hyperinsulinaemia and severe insulin resistance (ESM Fig. 4). Consistently, HFD feeding resulted in substantial weight gain (26.6±0.6 and 46.0±1.1 g in control [n=6] and HFD-fed [n=5] CMMR mice, respectively) and increased fasting blood glucose (3.1±0.2 and 6.0±0.4 mmol/l in control [n=6] and HFD-fed [n=5] CMMR mice, respectively). As shown in Fig. 2a,b, HFD feeding clearly increased the number of mitophagy signals in beta cells. The HFD-induced elevation of mitophagy signal was also observed in female mice (ESM Fig. 5). These results indicated that metabolic stress induced by HFD feeding enhanced mitophagy in beta cells.
HFD feeding elevated mitophagy in beta cells in HFD-G islets. (a) Representative images of pancreatic sections from CMMR mice fed a control diet or HFD for 20 weeks (green, EGFP; red, mCherry). (b) Relative numbers of mitophagy signals in pancreatic sections from CMMR mice fed a control diet or HFD for 20 weeks (n=45 and 55 islets for control and HFD, respectively). ***p<0.001, Mann–Whitney U test. (c) Control, HFD-SM and HFD-G islets were subjected to immunoblotting with an anti-mCherry antibody. (d) Relative degrading CMMR to full-length CMMR signal ratios (n=9 for each group). *p<0.05, paired t test, p values were adjusted by Holm’s method. (e) Control, HFD-SM and HFD-G islets were subjected to immunoblotting for LC3, p62 and VAMP7. Relative amounts of LC3-II (f: n=9 for each group), p62 (g: n=7 for each group) and VAMP7 (h: n=7 for each group). *p<0.05, paired t test, p values were adjusted by Holm’s method. (i) Representative images of control, HFD-SM and HFD-G islet sections. Islets were isolated from CMMR mice fed a control diet or HFD for 20 weeks to prepare frozen sections. (j) Relative numbers of mitophagy signals in isolated islets (n=51, 44 and 25 islets for control, HFD-SM and HFD-G, respectively). ***p<0.001, Mann–Whitney U test, p values were adjusted by Holm’s method. (k) Representative electron micrographs of control, HFD-SM and HFD-G islets treated with 30 nmol/l bafilomycin A1 for 24 h. Arrowheads indicate mitochondria located in autophagosomes. (l) Ratios of the number of mitochondria located in autophagosomes to the total number of mitochondria in each image (n=100 for each group). **p<0.01, ***p<0.001, Mann–Whitney U test, p values were adjusted by Holm’s method. Bars, 50 μm (a, i) and 2 μm (k)
To validate the upregulation of mitophagy in beta cells in metabolically stressed mice induced by HFD feeding, we evaluated mitophagy biochemically using the CMMR probe. During the degradation of the mCherry–EGFP probe in lysosomes, we previously demonstrated that EGFP was degraded faster than mCherry [26]. Accordingly, after the degradation of EGFP from the full-length CMMR probe, the consequent CMMR probe (degrading CMMR probe) is detected as an mCherry-only sequence. Indeed, an antibody against mCherry detected a major signal near 60 kDa corresponding to the molecular mass of the full-length CMMR probe and a minor signal around 30 kDa corresponding to the molecular mass of mCherry in islets from CMMR mice (Fig. 2c). Thus, the amount of the degrading CMMR probe detected around 30 kDa was evaluated to assess mitophagic activity in beta cells. We first examined size-matched islets with a diameter of 120–180 μm because these were the most abundant in isolated islets (ESM Fig. 6a). Although the amount of degrading CMMR probe was significantly increased in HFD-SM islets (Fig. 2c,d), the difference was smaller than expected based on the results shown in Fig. 2a,b. Because HFD feeding increases islet size [27], we next focused on enlarged islets with a diameter of >320 μm, which were characteristically increased in the pancreas of HFD-fed mice (ESM Fig. 6a). As shown in Fig. 2c,d, the amounts of the degrading CMMR probe were markedly elevated in HFD-G islets compared with that in control islets. We biochemically examined the autophagy marker microtubule-associated proteins 1A/1B light chain 3B (LC3) in islets. Autophagy converts the cytosolic form of LC3 (LC3-I) to lipidated LC3-II, which accumulates in autophagosomes and reflects the amount of autophagosomes [28]. As shown in Fig. 2e,f, the amount of LC3-II was markedly increased in HFD-G islets. Furthermore, p62/sequestosome 1 (SQSTM1, hereafter p62), which bridges general autophagic targets with autophagosomes and is consequently degraded in lysosomes [28], and vesicle-associated membrane protein 7 (VAMP7), which was shown to regulate mitophagy in beta cells [13, 22], were also increased in HFD-G islets (Fig. 2e,g,h). In contrast, the amounts of glucokinase and GLUT2 in HFD-SM and HFD-G islets were equivalent to those in control islets (ESM Fig. 7), suggesting that the expression of autophagy/mitophagy-related proteins was selectively upregulated in HFD-G islets.
To verify these results, we next examined the fluorescent mitophagy signals in islets isolated from control and HFD-fed CMMR mice. As expected from Fig. 2c,d, mitophagy signals were markedly elevated in HFD-G islets compared with those in control and HFD-SM islets (Fig. 2i,j). To further confirm these results, we examined endogenously induced mitophagy in isolated islets by electron microscopy. Control, HFD-SM and HFD-G islets were isolated and cultured for 24 h in the presence of bafilomycin A1. Using electron microscopy, we observed that the number of mitochondria in autophagosomes was significantly increased in HFD-SM islets and further increased in HFD-G islets (Fig. 2k,l). Collectively, these results demonstrated that HFD feeding activated mitophagy in beta cells, especially in HFD-G islets.
Aberrant accumulation of dysfunctional mitochondria in HFD-G islets
Increased mitophagy in HFD-G islets suggested that these islets contain a large amount of mitophagic targets. To address this point, we next examined endogenous ROS levels in cultured islet cells because mitochondrial dysfunction often leads to excessive ROS production [29]. Pancreatic islet cells cultured on coverslips were stained with MitoSOX, a mitochondrial superoxide indicator. As shown in Fig. 3a,b, the MitoSOX signal intensities in cells cultured from HFD-G islets were significantly higher than those from control and HFD-SM islets. The OxyBlot revealed that the amount of proteins oxidised by free radicals, such as ROS, was notably increased in HFD-G islets (Fig. 3c,d). Then, we visualised functional mitochondria using MTR (Fig. 3e,f). The signal intensities of MTR were significantly reduced in beta cells cultured from HFD-G islets, suggesting the aberrant accumulation of dysfunctional mitochondria. This accumulation of dysfunctional mitochondria may represent increased dysfunctional mitochondria and/or reduced degradation [28]. However, the significant increase in LC3-II induced by bafilomycin A1 treatment in HFD-G islets (ESM Fig. 8a, b) and the lysosome-dependent degradation signal of the mCherry–EGFP probe shown in Fig. 2c,d [26, 30] suggested that lysosomes in HFD-G islets were functional. Furthermore, the expression levels of p62 and transcription factor binding to IGHM enhancer 3 (TFE3), a master regulator of lysosomal genes [31], were elevated in HFD-G islets (ESM Fig. 8c, d). Considering the upregulation of mitophagy in HFD-G islets (Fig. 2), these results suggested that the accumulation of dysfunctional mitochondria is caused by an increase in targets to be degraded by autophagy, not a defect in lysosomal degradation. In addition, the generation of dysfunctional mitochondria would overwhelm the mitophagic capacity, resulting in the aberrant accumulation of dysfunctional mitochondria in HFD-G islets.
Accumulation of dysfunctional mitochondria in HFD-G islets. (a) Representative images of MitoSOX staining. Pancreatic islet cells cultured from control, HFD-SM and HFD-G islets were stained with 5 μmol/l MitoSOX for 20 min at 37°C. (b) Relative MitoSOX signal intensities (n=25, 27 and 30 images for control, HFD-SM and HFD-G, respectively). ***p<0.001, Mann–Whitney U test, p values were adjusted by Holm’s method. (c) Oxidised carbonyl groups in protein extracts from control, HFD-SM and HFD-G islets were labelled with 2,4-dinitrophenylhydrazine (DNPH) and then subjected to immunoblotting with an anti-2,4-dinitrophenyl (DNP) moiety antibody, using OxyBlot assays. (d) Relative OxyBlot signal intensities (n=7 for each group). Signal intensities of the OxyBlot from 35 to 140 kDa were quantified. *p<0.05, paired t test, p values were adjusted by Holm’s method. (e) Pancreatic beta cells cultured from control, HFD-SM and HFD-G islets were stained with 0.5 μmol/l MTR (red) for 30 min, followed by immunostaining for insulin (grey) and Tom20 (green). (f) Relative MTR/Tom20 signal ratios (n=30, 32 and 32 images for control, HFD-SM and HFD-G, respectively). **p<0.01, Tukey’s HSD test. Bars, 5 μm (a, e). HSD, honest significant difference
HFD-induced hypoxia upregulated BNIP3 expression through Hif-1α to enhance mitophagy in beta cells
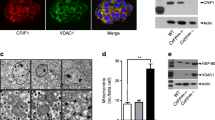
In various tissues, PINK1 is targeted to dysfunctional mitochondria, accumulates on their outer mitochondrial membrane (OMM) and recruits Parkin to initiate mitophagy [7, 8]. Thus, we hypothesised that the accumulation of dysfunctional mitochondria in HFD-G islets activates the PINK1/Parkin pathway to enhance mitophagy. Although we were unable to detect PINK1 in islets by immunoblotting, likely due to its weak expression [32] (data not shown), Parkin was significantly increased in HFD-SM and HFD-G islets (Fig. 4a,b). This result prompted us to examine the effect of Parkin overexpression on mitophagy in beta cells. However, the overexpression of haemagglutinin (HA)-tagged Parkin did not affect the mitophagy signal in CMMR beta cells (Fig. 4c,d). To examine the role of PINK1 in mitophagy in beta cells, CMMR mice were crossed with Pink1−/− mice to generate Cmmrflox/+;Cre;Pink1−/− mice. We found that the genetic ablation of PINK1 in beta cells did not affect mitophagy in CMMR mice fed a control diet (Fig. 4e,f) or HFD (Fig. 4g,h). In addition, Pink1 deletion did not affect glucose tolerance or GSIS (ESM Fig. 9). These results indicated that the upregulation of Parkin in beta cells did not affect mitophagy through the PINK1/Parkin pathway.
PINK1/Parkin pathway was not involved in mitophagy in pancreatic beta cells. (a) Control, HFD-SM and HFD-G islets were subjected to immunoblotting with specific antibodies as indicated. (b) Relative amounts of Parkin (n=6 for each group). *p<0.05, paired t test, p values were adjusted by Holm’s method. (c) CMMR beta cells expressing HA-Parkin were starved for 3 h, followed by immunostaining for HA-tag (blue). (d) Relative amounts of mitophagy signals (n=14 for each group). NS p>0.05, Mann–Whitney U test. (e, g) Representative images of pancreatic sections from 10-week-old Cmmrflox/+;Cre;Pink1+/+ (WT) and Cmmrflox/+;Cre;Pink−/− (PINK1 KO) mice (e) and from Cmmrflox/+;Cre;Pink1+/+ (WT + HFD) and Cmmrflox/+;Cre;Pink1−/− (PINK1 KO + HFD) mice fed the HFD for 20 weeks (g) (green, EGFP; red, mCherry). (f, h) Relative amounts of mitophagy signals (f: n=42 and 43 islets for WT and PINK1 KO, respectively; h: n=24, 41 and 40 islets for WT, WT + HFD and PINK1 KO + HFD, respectively). **p<0.01, NS p>0.1, Mann–Whitney U test, p values were adjusted by Holm’s method. Bars, 5 μm (c) and 50 μm (e, g). KO, knockout; mCh, mCherry; WT, wild type
Hypoxia induces mitophagy in various tissues [33, 34], and the islets of diabetic animals were shown to be exposed to hypoxic stress [3, 21]. As an alternative pathway to upregulate mitophagy, we studied hypoxia. Oxygen tension in islets was examined using pimonidazole, which forms adducts with intracellular molecules only in severe hypoxic conditions [21]. As shown in Fig. 5a,b, the number of pimonidazole adducts was moderately but significantly increased in HFD-SM islets and further increased in HFD-G islets. Exposure of cultured beta cells to hypoxic conditions (3% O2) for 24 h significantly increased ROS generation (Fig. 5c,d) and upregulated mitophagy (Fig. 5e,f). Consistent with the marked elevation in pimonidazole signals, the amount of Hif-1ɑ, a transcription factor stabilised and activated by hypoxia [35], was significantly increased in HFD-G islets. Furthermore, we found that the amounts of BNIP3 and BCL2 interacting protein 3 like (Nix/BNIP3L, hereafter, Nix), which are mitophagy receptors localised on the OMM that interact with LC3 to initiate mitophagy [7, 8], were also elevated in HFD-G islets (Fig. 5g–j). The hypoxia-induced upregulation of BNIP3 and Nix was mediated by Hif-1ɑ in Min6 cells and cultured beta cells (ESM Fig. 10), as reported in other cell types [7, 8]. Next, the effects of BNIP3 and Nix on mitophagy in beta cells were examined by overexpression and depletion experiments using CMMR beta cells. The overexpression of HA-tagged BNIP3 significantly increased mitophagy signals compared with TagRFP657 overexpression, whereas Nix overexpression did not affect mitophagy signals (Fig. 5k,l). Treatment with BNIP3 and Nix shRNA effectively reduced their targets (ESM Fig. 11). However, BNIP3 but not Nix shRNA significantly reduced mitophagy signals induced by DFP treatment (Fig. 5m,n). Finally, we examined the effect of BNIP3 and Hif-1ɑ shRNA on mitophagy induced by HFD. As shown in Fig. 5o,p, BNIP3 and Hif-1ɑ knockdown significantly suppressed mitophagy in cultured beta cells from HFD-G islets. These results indicated that hypoxia in HFD-G islets activated Hif-1ɑ to upregulate BNIP3, which subsequently enhanced mitophagy in beta cells.
Hif-1ɑ-mediated BNIP3 upregulation promoted mitophagy in pancreatic beta cells. (a) Control, HFD-SM and HFD-G islets were isolated from pimonidazole-injected control and HFD-fed mice and subjected to immunoblotting with an anti-pimonidazole antibody. (b) Relative pimonidazole signal intensities (n=5 for each group). The signal intensities of pimonidazole adducts from 50 to 140 kDa were quantified. **p<0.01, ***p<0.001, paired t test, p values were adjusted by Holm’s method. (c) Representative images of MitoSOX imaging. Pancreatic beta cells cultured under control and hypoxic conditions (3% O2) were stained with 5 μmol/l MitoSOX for 20 min at 37°C. (d) Relative MitoSOX signal intensities (n=27 for both control and hypoxia). ***p<0.001, Student’s t test. (e) Representative images of CMMR beta cells (green, EGFP; red, mCherry) cultured under control and hypoxic conditions. (f) Relative amounts of mitophagy signals (n=24 and 20 images for control and hypoxia, respectively). ***p<0.001, Welch’s t test. (g) Control, HFD-SM and HFD-G islets were subjected to immunoblotting with specific antibodies as indicated. (h–j) Relative amounts of Hif-1ɑ (h), Nix (i) and BNIP3 (j) (n=8 for each group). *p<0.05, paired t test, p values were adjusted by Holm’s method. (k) CMMR beta cells (green, EGFP; red, mCherry) expressing TagRFP657 (blue), HA-BNIP3 or HA-Nix were starved for 3 h, followed by immunostaining for HA-tag (blue). (l) Relative amounts of mitophagy signals (n=16, 15 and 16 images for TagRFP657, HA-BNIP3 and HA-Nix, respectively). ***p<0.001, Mann–Whitney U test, p values were adjusted by Holm’s method. (m) CMMR beta cells expressing control shRNA, BNIP3 shRNA or Nix shRNA were treated with 1 mmol/l DFP for 18 h. (n) Relative amounts of mitophagy signals (n=14, 11 and 15 for control shRNA, BNIP3 shRNA and Nix shRNA without DFP treatment, respectively; n=26, 15 and 20 for control shRNA, BNIP3 shRNA and Nix shRNA with DFP treatment, respectively). TagRFP657 was used as an infection marker. **p<0.01, Mann–Whitney U test, p values were adjusted by Holm’s method. (o) Control, BNIP3 or Hif-1ɑ shRNA was expressed in CMMR beta cells cultured from control or HFD-G islets. (p) Relative amounts of mitophagy signals (n=21 for control beta cells expressing control shRNA, and n=20, 16 and 18 for HFD-G beta cells expressing control shRNA, BNIP3 shRNA and Hif-1a shRNA, respectively). TagRFP657 was used as an infection marker. **p<0.01, Tukey’s HSD test. Bars, 5 μm (c, e, k, m, o). Ctrl, control; HSD, honest significant difference; Hypo, hypoxia; Pimo, pimonidazole
Accumulation of dysfunctional mitochondria affected mitochondrial function and insulin secretion in pancreatic beta cells
The accumulation of dysfunctional mitochondria exacerbates mitochondrial function [29]. Thus, we next examined the effect of the aberrant accumulation of dysfunctional mitochondria on mitochondrial function and insulin secretion. Mitochondrial function in isolated islets was evaluated as the OCR. As shown in Fig. 6a,b, the OCR in response to glucose stimulation was markedly reduced in HFD-G islets. Although the amount of secreted insulin in response to glucose stimulation was higher in HFD-G islets (103±4, 132±12 and 444±39 fmol/islet for control, HFD-SM and HFD-G islets stimulated with 16 mmol/l glucose, respectively), the efficiency of insulin secretion, represented by the ratio of secreted insulin to total insulin content, was significantly decreased in HFD-G islets (Fig. 6c). In contrast, we found that the OCR and insulin secretion were significantly enhanced in HFD-SM islets.
Chronic administration of insulin reduced mitophagy and restored mitochondrial function and insulin secretion in beta cells from HFD-fed mice. (a) The OCRs in control, HFD-SM and HFD-G islets were measured by a flux analyser. Values were normalised to basal respiration (% of baseline) (n=10, 10 and 13 for control, HFD-SM and HFD-G, respectively). (b) AUC analysis of (a). **p<0.01, ***p<0.001, Mann–Whitney U test, p values were adjusted by Holm’s method. (c) Control, HFD-SM and HFD-G islets were stimulated with 2.2 or 16 mmol/l glucose for 30 min (n=4 for control, HFD-SM and HFD-G islets stimulated with 2.2 mmol/l glucose; n=6, 6 and 7 for control, HFD-SM and HFD-G stimulated with 16 mmol/l glucose, respectively). *p<0.05, **p<0.01, Tukey’s HSD test. (d) HFD-fed mice were chronically treated with saline or insulin for 6 weeks. Pimonidazole was injected into these mice, and then HFD-SM and HFD-G islets were isolated and subjected to immunoblotting with an anti-pimonidazole antibody. (e) Relative pimonidazole signal intensities (n=8 for each group). Signal intensities of pimonidazole adducts from 50 to 150 kDa were quantified. *p<0.05, **p<0.01, paired t test, p values were adjusted by Holm’s method. (f) Representative images of pancreatic sections from CMMR mice fed a control diet or HFD for 20 weeks (green, EGFP; red, mCherry). Insulin or saline was chronically administered to HFD-fed CMMR mice for the last 6 weeks. (g) Relative amounts of mitophagy signals (n=23, 32 and 36 islets for control, HFD + Saline and HFD + Ins, respectively). ***p<0.001, Welch’s t test, p values were adjusted by Holm’s method. (h) Representative images of MitoSOX staining. Pancreatic islet cells cultured from saline-treated HFD-SM and HFD-G islets and insulin-treated HFD-SM and HFD-G islets were stained with 5 μmol/l MitoSOX for 20 min at 37°C. (i) Relative MitoSOX signal intensities (n=26, 33, 24 and 28 images for saline-treated HFD-SM, insulin-treated HFD-SM, saline-treated HFD-G and insulin-treated HFD-G, respectively). **p<0.01, Tukey’s HSD test. (j) Pancreatic beta cells cultured from saline-treated HFD-SM and HFD-G islets and insulin-treated HFD-SM and HFD-G islets were stained with 0.5 μmol/l MTR (red) for 30 min, followed by immunostaining for insulin (grey) and Tom20 (green). (k) Relative MTR/Tom20 signal ratios (n=24, 22, 24 and 24 images for saline-treated HFD-SM, insulin-treated HFD-SM, saline-treated HFD-G and insulin-treated HFD-G, respectively). ***p<0.001, Welch’s t test, p values were adjusted by Holm’s method. (l) Saline-treated HFD-SM and HFD-G islets and insulin-treated HFD-SM and HFD-G islets were stimulated with 16 mmol/l glucose for 30 min (n=12, 12, 5 and 5 for saline-treated HFD-SM, insulin-treated HFD-SM, saline-treated HFD-G and insulin-treated HFD-G, respectively). **p<0.01, Tukey’s HSD test. Bars, 20 μm (f) and 5 μm (h, j). AU, arbitrary units; HSD, honest significant difference; Ins, insulin; Pimo, pimonidazole; Rot/AA, rotenone/antimycin A
In contrast to HFD-G islets, dysfunctional mitochondria did not accumulate in HFD-SM islets (Fig. 3). Thus, we hypothesised that the accumulation of dysfunctional mitochondria might impair mitochondrial function and insulin secretion in HFD-G islets. To test this hypothesis, we examined whether the accumulation of dysfunctional mitochondria could be reduced by chronic insulin treatment, a well-known therapy to preserve functional beta cells in type 2 diabetic patients [36]. Mice fed the HFD for 20 weeks were chronically treated with insulin for the last 6 weeks (HFD-Ins mice), which significantly improved glucose tolerance (ESM Fig. 12). As shown in Fig. 6d,e, insulin treatment significantly reduced the amount of pimonidazole adducts in HFD-G islets, suggesting that insulin reduced the oxygen consumption required to secrete large amounts of insulin in beta cells [21]. Consistently, chronic insulin treatment significantly reduced mitophagy signals in beta cells in HFD-Ins mice (Fig. 6f,g). Next, to assess the number of dysfunctional mitochondria in beta cells in HFD-Ins mice, beta cells were cultured from HFD-SM and HFD-G islets and stained with MitoSOX to visualise ROS production. As shown in Fig. 6h,i, chronic insulin treatment reduced ROS production in beta cells cultured from HFD-SM and HFD-G islets. We also found that MTR signals in beta cells cultured from control-treated HFD-G islets were decreased compared with those cultured from control-treated HFD-SM islets, and were restored by chronic insulin treatment (Fig. 6j,k). Finally, we found that insulin secretion from HFD-SM and HFD-G islets was improved by chronic insulin treatment (Fig. 6l). Collectively, these results demonstrated that the aberrant accumulation of dysfunctional mitochondria resulted in impaired insulin secretion in HFD-G islets.
Discussion
In this study, we generated CMMR mice to evaluate mitophagy in beta cells. We showed that mitophagy was upregulated in beta cells through the hypoxia/Hif-1ɑ/BNIP3 axis in HFD-induced diabetic model mice. Moreover, insufficient mitophagy relative to the substantially increased generation of dysfunctional mitochondria caused the accumulation of dysfunctional mitochondria, defective maintenance of mitochondrial function and impaired insulin secretion, especially in HFD-G islets.
Previously, two mitophagy reporter mouse lines have been reported: the mt-Keima mouse line expressing the pH-sensitive fluorescence resonance energy transfer (FRET) probe [14] and the mito-QC mouse line expressing tandemly ligated mCherry–EGFP [15, 19]. However, mt-Keima mice were not applicable for evaluating mitophagy in vivo because the probe was incompatible with fixation [15]. Moreover, beta cells in mito-QC mice have been used to examine mitophagy by FACS [37], but the fluorescent signals in their pancreatic islets were too weak to detect mitophagy in vivo [19]. Although both CMMR and mito-QC probes are based on tandemly ligated mCherry–EGFP, the fluorescent probe is expressed in the mitochondrial matrix in our CMMR mice but on the OMM in mito-QC mice. Thus, the localisation of mCherry–EGFP likely affects its signal intensity in pancreatic beta cells. In addition, our results suggested that the expression of the CMMR probe did not disrupt mitochondrial function in pancreatic beta cells (ESM Fig. 1). Furthermore, our results (Fig. 1) and cumulative evidence [18, 19, 28] demonstrated that tandemly ligated mCherry–EGFP convincingly detected a change in the pH environment during autophagy/mitophagy. Thus, our CMMR probe serves as a reliable marker for mitophagy in beta cells.
In this study, mitophagy was upregulated in beta cells in HFD feeding-induced diabetic model mice. Because these mice exhibited hyperinsulinaemia (ESM Fig. 4), mitochondria in beta cells likely increase ATP production to secrete large amounts of insulin, generating excessive ROS and dysfunctional mitochondria. Thus, the upregulation of mitophagy observed in beta cells in HFD-fed mice may represent an adaptive response to degrade dysfunctional mitochondria. In this situation, the balance between the generation and degradation of dysfunctional mitochondria determines the response to glucose stimulation. In HFD-SM islets, maintenance of mitochondrial function by reasonably upregulated mitophagy would support active insulin secretion. Conversely, overwhelming the mitophagic capacity by excessive generation of dysfunctional mitochondria leads to defective maintenance of mitochondrial function and impairment of insulin secretion in HFD-G islets. We also note that not only dysfunctional mitochondria but also other autophagic targets to be degraded should accumulate in HFD-G islets, and may cause the beta cell dysfunction. Consistent with our results using HFD-SM and HFD-G islets, previous studies showed that diabetes increased the number of enlarged islets with a reduced insulin secretion capacity compared with smaller islets [27, 38,39,40]. We also assumed that the functional difference between HFD-SM and HFD-G islets resulted from the different durations of hyper-insulin secretion. Compared with HFD-SM islets, HFD-G islets are expected to experience a longer period of hyper-insulin secretion, leading to the chronic generation of excessive ROS and dysfunctional mitochondria. In fact, alleviating hyper-insulin secretion by chronic insulin treatment reduced ROS production and mitophagy signals and restored insulin secretion in HFD-G islets (Fig. 6), similar to type 2 diabetes patients [36]. Notably, how insulin treatment alleviated maintenance of mitochondrial function through mitophagy in beta cells remains unclear. It would be interesting to examine whether a reduction of the blood glucose level or signalling process through the insulin receptor could restore the gross function of mitochondria and the beta cell function in the future. Our results showed that HFD-G islets secreted a large amount of insulin, but their beta cells accumulated dysfunctional mitochondria, consequently leading to impaired insulin secretion. Although HFD-G islets accounted for 10.3±2.6% of the total number of islets, this mathematically corresponded to 44.8±9.2% of the total islet volume (ESM Fig. 6), suggesting that beta cell dysfunction in HFD-G islets would have a certain impact on the pathogenesis of type 2 diabetes. However, further studies are required to address this point.
In this study, we showed that BNIP3 mediated mitophagy in beta cells (Fig. 5). Consistent with previous reports [41, 42], the expression of BNIP3 depended on Hif-1ɑ and facilitated mitophagy in beta cells. Thus, it is reasonable to suggest that chronic hyper-insulin secretion induced severe hypoxia, which subsequently upregulated BNIP3 via Hif-1ɑ to promote mitophagy as an adaptive response in HFD-G islets. Although PINK1/Parkin is a well-known pathway that regulates mitophagy, we did not find that it mediated mitophagy in beta cells. However, we cannot rule out the possible involvement of Parkin in mitophagy in beta cells because some studies suggested that a PINK1-independent but Parkin-mediated mechanism was implicated in mitophagy [43, 44].
In conclusion, we generated a powerful tool to evaluate mitophagy in pancreatic beta cells. We also demonstrated that metabolic stress induced by HFD feeding elevated mitophagy in beta cells, likely through the upregulation of BNIP3 via cellular hypoxia and Hif-1ɑ. An imbalance between the generation and degradation of dysfunctional mitochondria reduced mitochondrial function and impaired insulin secretion. Further studies are necessary to elucidate the molecular mechanism of mitophagy in pancreatic beta cells.
Data availability
The datasets generated and/or analysed during this study are available from the corresponding author upon reasonable request.
Abbreviations
- Atg7:
-
Autophagy related 7
- BNIP3:
-
BCL2 interacting protein 3
- CQ:
-
Chloroquine
- CMMR:
-
Conditional mitochondrial matrix targeting mitophagy reporter
- DFP:
-
Deferiprone
- EGFP:
-
Enhanced GFP
- GSIS:
-
Glucose-stimulated insulin secretion
- HA:
-
Haemagglutinin
- HFD:
-
High-fat diet
- HFD-G:
-
Enlarged islets from HFD-fed mice
- HFD-Ins mice:
-
Mice fed HFD for 20 weeks then treated with insulin for 6 weeks
- HFD-SM:
-
Size-matched islets from HFD-fed mice
- Hif-1ɑ:
-
Hypoxia-inducible factor 1-ɑ
- LAMP1:
-
Lysosomal-associated membrane protein 1
- LC3:
-
Microtubule-associated proteins 1A/1B light chain 3B
- LC3-I:
-
Cytosolic form of LC3
- LC3-II:
-
Lipidated form of LC3
- MTR:
-
MitoTracker Red CM-H2Xros
- Nix/BNIP3L:
-
BCL2 interacting protein 3 like
- OCR:
-
Oxygen consumption rate
- OMM:
-
Outer mitochondrial membrane
- PINK1:
-
PTEN-induced kinase 1
- ROS:
-
Reactive oxygen species
- shRNA:
-
Short hairpin RNA
- Tom20:
-
Translocase of outer mitochondrial membrane 20
- VAMP7:
-
Vesicle-associated membrane protein 7
References
Hudish LI, Reusch JE, Sussel L (2019) β cell dysfunction during progression of metabolic syndrome to type 2 diabetes. J Clin Invest 129(10):4001–4008. https://doi.org/10.1172/JCI129188
Yagihashi S, Inaba W, Mizukami H (2016) Dynamic pathology of islet endocrine cells in type 2 diabetes: β-cell growth, death, regeneration and their clinical implications. J Diabetes Investig 7(2):155–165. https://doi.org/10.1111/jdi.12424
Gerber PA, Rutter GA (2017) The role of oxidative stress and hypoxia in pancreatic beta-cell dysfunction in diabetes mellitus. Antioxid Redox Signal 26(10):501–518. https://doi.org/10.1089/ars.2016.6755
Chen C, Cohrs CM, Stertmann J, Bozsak R, Speier S (2017) Human beta cell mass and function in diabetes: Recent advances in knowledge and technologies to understand disease pathogenesis. Mol Metab 6(9):943–957. https://doi.org/10.1016/j.molmet.2017.06.019
Ashcroft FM, Rorsman P (2012) Diabetes mellitus and the β cell: the last ten years. Cell 148(6):1160–1171. https://doi.org/10.1016/j.cell.2012.02.010
Las G, Oliveira MF, Shirihai OS (2020) Emerging roles of β-cell mitochondria in type-2-diabetes. Mol Asp Med 71:100843. https://doi.org/10.1016/j.mam.2019.100843
Killackey SA, Philpott DJ, Girardin SE (2020) Mitophagy pathways in health and disease. J Cell Biol 219(11):e202004029. https://doi.org/10.1083/jcb.202004029
Palikaras K, Lionaki E, Tavernarakis N (2018) Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol 20(9):1013–1022. https://doi.org/10.1038/s41556-018-0176-2
Jung HS, Chung KW, Won Kim J et al (2008) Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab 8(4):318–324. https://doi.org/10.1016/j.cmet.2008.08.013
Ebato C, Uchida T, Arakawa M et al (2008) Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 8(4):325–332. https://doi.org/10.1016/j.cmet.2008.08.009
Soleimanpour SA, Gupta A, Bakay M et al (2014) The diabetes susceptibility gene Clec16a regulates mitophagy. Cell 157(7):1577–1590. https://doi.org/10.1016/j.cell.2014.05.016
Bartolomé A, Kimura-Koyanagi M, Asahara S et al (2014) Pancreatic β-cell failure mediated by mTORC1 hyperactivity and autophagic impairment. Diabetes 63(9):2996–3008. https://doi.org/10.2337/db13-0970
Aoyagi K, Ohara-Imaizumi M, Itakura M et al (2016) VAMP7 regulates autophagy to maintain mitochondrial homeostasis and to control insulin secretion in pancreatic β-cells. Diabetes 65(6):1648–1659. https://doi.org/10.2337/db15-1207
Sun N, Yun J, Liu J et al (2015) Measuring in vivo mitophagy. Mol Cell 60(4):685–696. https://doi.org/10.1016/j.molcel.2015.10.009
McWilliams TG, Prescott AR, Allen GF et al (2016) mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol 214(3):333–345. https://doi.org/10.1083/jcb.201603039
Katayama H, Hama H, Nagasawa K et al (2020) Visualizing and modulating mitophagy for therapeutic studies of neurodegeneration. Cell 181(5):1176–1187. https://doi.org/10.1016/j.cell.2020.04.025
Varuzhanyan G, Ladinsky MS, Yamashita SI et al (2021) Fis1 ablation in the male germline disrupts mitochondrial morphology and mitophagy, and arrests spermatid maturation. Development 148(16):dev199686. https://doi.org/10.1242/dev.199686
Allen GF, Toth R, James J, Ganley IG (2013) Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep 14(12):1127–1135. https://doi.org/10.1038/embor.2013.168
McWilliams TG, Prescott AR, Montava-Garriga L et al (2018) Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab 27(2):439–449. https://doi.org/10.1016/j.cmet.2017.12.008
Kulkarni RN, Brüning JC, Winnay JN et al (1999) Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96(3):329–339. https://doi.org/10.1016/s0092-8674(00)80546-2
Sato Y, Endo H, Okuyama H et al (2011) Cellular hypoxia of pancreatic β-cells due to high levels of oxygen consumption for insulin secretion in vitro. J Biol Chem 286(14):12524–12532. https://doi.org/10.1074/jbc.m110.194738
Aoyagi K, Itakura M, Fukutomi T et al (2018) VAMP7 regulates autophagosomes formation by supporting Atg9 functions in pancreatic β-cells from male mice. Endocrinology 159(11):3674–3688. https://doi.org/10.1210/en.2018-00447
Ohara-Imaizumi M, Aoyagi K, Yamauchi H et al (2019) ELKS/Voltage-dependent Ca2+ channel-β subunit module regulates polarized Ca2+ influx in pancreatic β cells. Cell Rep 26(5):1213–1226. https://doi.org/10.1016/j.celrep.2018.12.106
Yamashita SI, Jin X, Furukawa K et al (2016) Mitochondrial division occurs concurrently with autophagosomes formation but independently of Drp1 during mitophagy. J Cell Biol 215(5):649–665. https://doi.org/10.1083/jcb.201605093
Chu KY, O'Reilly L, Ramm G, Biden TJ (2015) High-fat diet increases autophagic flux in pancreatic beta cells in vivo and ex vivo in mice. Diabetologia 58(9):2074–2078. https://doi.org/10.1007/s00125-015-3665-x
Yamashita SI, Kyuuma M, Inoue K et al (2021) Mitophagy reporter mouse analysis reveals increased mitophagy activity in disuse-induced muscle atrophy. J Cell Physiol 236(11):7612–7624. https://doi.org/10.1002/jcp.30404
Gupta D, Jetton TL, LaRock K et al (2017) Temporal characterization of β cell-adaptive and -maladaptive mechanisms during chronic high-fat feeding in C57BL/6NTac mice. J Biol Chem 292(30):12449–12459. https://doi.org/10.1074/jbc.m117.781047
Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140(3):313–326. https://doi.org/10.1016/j.cell.2010.01.028
Fischer F, Hamann A, Osiewacz HD (2012) Mitochondrial quality control: an integrated network of pathways. Trends Biochem Sci 37(7):284–292. https://doi.org/10.1016/j.tibs.2012.02.004
Katayama H, Yamamoto A, Mizushima N, Yoshimori T, Miyawaki A (2008) GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct 33(1):1–12. https://doi.org/10.1247/csf.07011
Raben N, Puertollano R (2016) TFEB and TFE3: Linking lysosomes to cellular adaptation to stress. Annu Rev Cell Dev Biol 32:255–278. https://doi.org/10.1146/annurev-cellbio-111315-125407
Deas E, Piipari K, Machhada A et al (2014) PINK1 deficiency in β-cells increases basal insulin secretion and improves glucose tolerance in mice. Open Biol 4(5):140051. https://doi.org/10.1098/rsob.140051
Chourasia AH, Boland ML, Macleod KF (2015) Mitophagy and cancer. Cancer Metab 3:4. https://doi.org/10.1186/s40170-015-0130-8
Wei H, Liu L, Chen Q (2015) Selective removal of mitochondria via mitophagy: distinct pathways for different mitochondrial stress. Biochim Biophys Acta 1853(10 Pt B):2784–2790. https://doi.org/10.1016/j.bbamcr.2015.03.013
Gunton JE (2020) Hypoxia-inducible factors and diabetes. J Clin Invest 130(10):5063–5073. https://doi.org/10.1172/jci137556
Swisa A, Glaser B, Dor Y (2017) Metabolic stress and compromised identity of pancreatic beta cells. Front Genet 8:21. https://doi.org/10.3389/fgene.2017.00021
Sidarala V, Pearson GL, Parekh VS et al (2020) Mitophagy protects β cells from inflammatory damage in diabetes. JCI insight 5(24):e141138. https://doi.org/10.1172/jci.insight.141138
Hayami T, Yokoi N, Yamaguchi T et al (2020) Tumor-like features of gene expression and metabolic profiles in enlarged pancreatic islets are associated with impaired incretin-induced insulin secretion in obese diabetes: A study of Zuker fatty diabetes mellitus rat. J Diabetes Investig 11(6):1434–1447. https://doi.org/10.1111/jdi.13272
Chan CB, Wright GM, Wadowska DW, MacPhail RM, Ireland WP, Sulston KW (1998) Ultrastructural and secretory heterogeneity of fa/fa (Zucker) rat islets. Mol Cell Endocrinol 136(2):119–129. https://doi.org/10.1016/s0303-7207(97)00220-7
MacGregor RR, Williams SJ, Tong PY, Kover K, Moore WV, Stehno-Bittel L (2006) Small rat islets are superior to large islets in in vitro function and in transplantation outcomes. Am J Physiol Endocrinol Metab 290(5):E771–E779. https://doi.org/10.1152/ajpendo.00097.2005
Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB (2012) Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem 287(23):19094–19104. https://doi.org/10.1074/jbc.m111.322933
Bellot G, Garcia-Medina R, Gounon P et al (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29(10):2570–2581. https://doi.org/10.1128/mcb.00166-09
Hoshino A, Ariyoshi M, Okawa Y et al (2009) Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic β-cell function in diabetes. Proc Natl Acad Sci USA 111(8):3116–3121. https://doi.org/10.1073/pnas.1318951111
Chen L, Liu C, Gao J et al (2017) Inhibition of Miro disturbs mitophagy and pancreatic β-cell function interfering insulin release via IRS-Akt-Foxo1 in diabetes. Oncotarget 8(53):90693–90705. https://doi.org/10.18632/oncotarget.20963
Acknowledgements
The authors thank R. Kulkarni (Joslin Diabetes Center, MA, USA) and K. Ueki (National Center for Global Health and Medicine, Tokyo, Japan) for the RIP-Cre mice, and S. Matsubara and J. Hayakawa (Laboratory for Electron Microscopy, Kyorin University School of Medicine, Tokyo, Japan) for their great technical contribution to electron microscopic analysis.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
KA, KS, TK and MO-I designed the study. KA, SY, YA, CN, YN and MA performed experiments. KA, SY, YA, HU, MA, KS, TK and MO-I analysed data. KA and MO-I wrote the original manuscript. All authors revised the manuscript critically for valuable intellectual content and approved the final version of the manuscript. MO-I is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology in Japan/Japan Society for the Promotion of Science KAKENHI (grant nos. 17K09845, 20K11563 [to KA], 17K08547, 21H02431 [to MO-I], 19H05712 [to TK]), a Junior Scientist Development grant supported by Novo Nordisk Pharma Ltd (to KA), The Naito Foundation (to MO-I) and AMED grant no. JP21gm6110013h004 (to TK).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM
(PDF 0.98 kb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aoyagi, K., Yamashita, Si., Akimoto, Y. et al. A new beta cell-specific mitophagy reporter mouse shows that metabolic stress leads to accumulation of dysfunctional mitochondria despite increased mitophagy. Diabetologia 66, 147–162 (2023). https://doi.org/10.1007/s00125-022-05800-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-022-05800-8