Abstract
Key Message
Lack of function of a D-genome adult plant resistance gene upon introgression into durum wheat.
Abstract
The wheat Lr34/Yr18/Sr57/Pm38/Ltn1 adult plant resistance gene (Lr34), located on chromosome arm 7DS, provides broad spectrum, partial, adult plant resistance to leaf rust, stripe rust, stem rust and powdery mildew. It has been used extensively in hexaploid bread wheat (AABBDD) and conferred durable resistance for many decades. These same diseases also occur on cultivated tetraploid durum wheat and emmer wheat but transfer of D genome sequences to those subspecies is restricted due to very limited intergenomic recombination. Herein we have introgressed the Lr34 gene into chromosome 7A of durum wheat. Durum chromosome substitution line Langdon 7D(7A) was crossed to Cappelli ph1c, a mutant derivative of durum cultivar Cappelli homozygous for a deletion of the chromosome pairing locus Ph1. Screening of BC1F2 plants and their progeny by KASP and PCR markers, 90 K SNP genotyping and cytology identified 7A chromosomes containing small chromosome 7D fragments encoding Lr34. However, in contrast to previous transgenesis experiments in durum wheat, resistance to wheat stripe rust was not observed in either Cappelli/Langdon 7D(7A) or Bansi durum plants carrying this Lr34 encoding segment due to low levels of Lr34 gene expression. Key message
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diseases known as rusts remain a constant threat to global production of both hexaploid bread wheat (Triticum aestivum L., 2n = 6x = 42) and tetraploid durum and cultivated emmer wheat (Triticum turgidum subsp. durum (Desf) Husn. and T. turgidum subsp. dicoccum (Schrank ex Schübl.) Thell., 2n = 4x = 28) (Kolmer 2005, 2015; Singh et al. 2011; Huerta-Espino et al. 2011; Bhavani et al. 2019). Two forms of resistance are commonly employed to help protect these crops from rusts, all stage resistance (ASR) and adult plant resistance (APR) (Singh et al. 2015; Ayliffe et al. 2022). ASR, also known as seedling resistance, can provide high levels of resistance but is generally only effective against a single pathogen species and usually race specific. While individual ASR genes can be very effective, they generally suffer from a lack of durability and are often rapidly overcome by pathogen mutation (Ayliffe et al. 2008).
In contrast, some (but not all) APR genes provide broad spectrum resistance that is generally partial and, in some instances, effective against multiple pathogen species. One such examples is the Lr34/Yr18/Sr57/Pm38/Ltn1 APR gene (hereafter referred to as Lr34) which has provided broad spectrum, partial resistance to leaf rust, stripe rust, stem rust and powdery mildew (Dyck et al. 1966; Kolmer et al. 2008; Herrera-Foessel et al. 2014). Lr34 encodes an ABC transporter (Krattinger et al. 2009) that is suggested to transport abscisic acid (Krattinger et al. 2019), although the exact mechanistic basis of resistance remains unclear. A remarkable feature of Lr34 is its durability (Dyck et al. 1966; Kolmer et al. 2008). It has been used for many decades in many wheat cultivars and remains durable, despite large scale deployment and constant rust pathogen evolution that continues to overcome many ASR genes (Singh et al. 2011; Olivera et al. 2015; Patpour et al. 2017).
Lr34 is encoded on chromosome arm 7DS of bread wheat thereby making this valuable gene unavailable for deployment in tetraploid wheat by standard conventional breeding. Previously Lr34 was introduced into durum wheat cultivar Stewart by Agrobacterium-mediated transformation (Rinaldo et al. 2017). Interestingly, these transgenic plants showed seedling resistance to rust diseases at ambient temperature which is atypical for this APR gene. Those transgenic durum plants with the highest levels of Lr34 expression showed the greatest resistance (Rinaldo et al. 2017). RNA analysis also showed that these resistant seedlings had elevated Lr34 expression compared with wild type bread wheat seedlings carrying Lr34. This increased expression was ascribed to the different genomic locations in which the transgene was integrated. These data confirmed that, with appropriate expression levels, Lr34 is functional in durum cultivar Stewart and no additional genes encoded on the wheat D genome are required for this seedling resistance. It is noteworthy that Lr34 is also functional in heterologous hosts such as maize, rice, barley and sorghum, albeit with pleiotropic effects associated with higher expression levels (Risk et al. 2013; Krattinger et al. 2016; Schnippenkoetter et al. 2017; Sucher et al. 2017).
In this study, we attempted to make the Lr34 gene available for durum breeding using a nonGM deployment strategy. A mutation in the Ph1 locus (ph1c) was used to promote homoeologous recombination (Sears 1977; Giorgi and Barbera 1981) in durum wheat between chromosome 7A and a 7D chromosome encoding Lr34. While this was successfully achieved, in contrast to previous transgenic experiments (Rinaldo et al. 2017), Lr34 resistance was not observed in plants carrying Lr34 on 7A/7D chromosomes. Expression analysis of field grown materials showed that Lr34 expression in these durum plants was significantly reduced when compared with bread wheat controls. Given the clear correlation between Lr34 resistance and gene expression levels observed in transgenic durum wheat (Rinaldo et al. 2017), this reduced expression likely explains the inability of this gene to provide resistance in this heterologous chromosome context.
Materials and methods
Germplasm
A durum wheat disomic substitution line containing a chromosome 7D substitution for chromosome 7A in cultivar Langdon (LDN), LDN 7D(7A) (Joppa and Williams 1988) was used as a parent. Chromosome 7D in this line was derived from bread wheat cultivar Chinese Spring (CS) which carries Lr34. The second parent was Cappelli ph1c, a durum wheat line that is homozygous for a deletion of the Ph1 locus (Giorgi and Barberra 1981). Cappelli/LDN 7D(7A) derived plants containing small 7D introgressions encoding Lr34 were tested for stripe rust resistance in field trials. These Cappelli/LDN 7D(7A) plants were also crossed for three generations using Bansi, an Indian landrace that is highly susceptible to Australian Puccinia striiformis f. sp. tritici Eriks. and Henn. (Pst) isolates, as a recurrent parent with homozygous derivatives of this material named Bansi Lr34. They were also backcrossed for six generations to Italian cultivar Svevo and homozygous progeny lines selected from selfed BC6 individuals to create Svevo Lr34. Both Bansi Lr34 and Svevo Lr34 was field trialled for resistance to stripe rust.
Plant growth conditions
Glasshouse plants were grown with a 16 h, 22 °C/8 h, 18 °C growth temperature regime in 25 cm pots containing a compost/soil mixture and fertilised by periodic application of osmocote slow-release fertiliser (Scotts, Australia). Plants grown in growth cabinets had a 16-h light/8-h dark photoperiod and 22 °C/16 °C temperature cycle. For speed breeding applications plants were grown in 15 cm pots in growth cabinets under a 22-h light/2-h dark photoperiod at a constant 22 °C. Speed breeding was used to backcross Lr34 introgressions from LDN 7D(7A)/Cappelli progeny to both Bansi and Svevo.
Field trials
Field trials were undertaken at the experimental sites, Horse Research Unit and Lansdowne, of the Plant Breeding Institute, University of Sydney, Cobbitty, NSW in 2021 and 2022, respectively. Plants were grown in replicated 1-m rows with seeds sowed 10 cm part. A mixture of susceptible cultivars (spreader) was planted every six rows to facilitate disease development and spread. Susceptible pots from the glasshouse were put in the field to initiate the rust development. Plants were infected at the 5-leaf stage with a mixed inoculum, consisting of predominant local Pst pathotypes. Experimental material was infected naturally throughout the growing season from uredinospores on adjacent spreader rows. Flag leaves were scored for adult plant resistance when the susceptible cultivar Bansi showed an 8–9 response on a 1–9 (very resistant to very susceptible) scale (Bariana et al. 2007).
Quantification of fungal biomass
Flag leaves (> 5) from plants grown in field disease nurseries were harvested just prior to senescence, weighed and processed as described by Ayliffe et al. (2013). Briefly, samples were autoclaved in 1 M KOH and then neutralised in 50 mM Tris pH 7.5. Samples were then homogenised with a sonicator and resuspended at 200 mg/ml in Tris buffer. Aliquots (200 μl) were stained with 10 μg of wheat germ agglutinin FITC for 30 min, washed 3 times and finally resuspended in 50 mM Tris buffer. Fluorometric measurements were made with a Wallac Victor 1420 multilabel counter (Perkin-Elmer Life Science, Waltham, MA, USA) fluorometer with 485-nm adsorption and 535-nm emission wavelengths and a 1.0-s measurement time. Triplicate reactions were undertaken for each sample.
DNA extraction for PCR analyses
Plant DNA was extracted from freeze dried tissue using a Nimbus liquid handling robot (Hamilton, Province, USA). Freeze dried leaf tissue was macerated with a ball bearing and heat treated for 1 h in extraction buffer (0.1 M Tris pH 8.0, 0.05 M EDTA, 1.25% SDS). After the addition of 6 M NH3Ac and centrifugation the supernatant was precipitated with isopropanol and DNA washed in 70% ethanol and finally resuspended in TE buffer (10 mM Tris pH 8.0, 1 mM EDTA).
DNA quantitative PCR analysis
DNA samples for quantitative DNA analysis of the wheat D genome specific repeat sequence, Dgas44 (Bryan et al. 1998; McNeil et al. 1994; Han et al. 2014), were amplified with GoTaq DNA polymerase (Promega) using a CFX96 Real Time System and C100 Touch Thermo Cycler (Bio‐Rad). Dgas44 amplification products were normalized relative to wheat 28S rRNA genes (Han et al. 2016; GenBank AY049041.1). The comparative CT method was used for quantification (Schmittgen and Livak 2008) with three replicate reactions per sample. Dgas44 and 28S rRNA specific primers are listed in Table S1.
KASP analysis of single-nucleotide and presence–absence polymorphisms
Six single-nucleotide polymorphisms (SNPs) between sequences on chromosome 7D of bread wheat and the homeologous sequences on chromosome 7A of durum wheat were assayed on 71 progeny plants using Kompetitive Allele-specific PCR (KASP; LGC/Biosearch, UK) primer sets (Table S1). Genotyping was performed on a SNPline platform (LGC/Biosearch, UK), according to the manufacturer’s instructions. Similarly, KASP primer sets (Table S1) were used to detect the presence of the chromosome-7D bread wheat genes Lr34 (primer set Lr34_TCCIND, Rasheed et al. 2016) and Xat (GenBank MN233788; Watkins et al. 2019), neither of which is present in durum wheat.
Gel-based analysis of presence–absence and length polymorphisms
Chromosome 7D markers were generated by comparing random regions of CS chromosome 7D sequence to chromosome 7A to identify potential deletion polymorphisms. Primers were then designed to either amplify products only from chromosome 7D or amplify products of different lengths from chromosomes 7D and 7A. PCR products were amplified from CS using a Phire Plant Direct PCR Kit (Thermo Scientific, USA), resolved by 1% agarose gel electrophoresis and sequenced by Sanger sequencing to confirm their identities. Primer pairs were then screened against LDN and LDN 7D(7A) to identify 7D-specific markers. Four markers (Cs7D1, Cs7D2, Cs7D3 and Cs7D4) identified by this approach (Table S1) were selected for genotyping of progeny plants. The presence of Lr34 in germplasm was also determined using dominant gene specific PCR amplification primers, ABCTF4N (Krattinger et al. 2009) and Lr34 PLUSR (Lagudah et al. 2009), derived from the cloned Lr34 gene sequence.
Array-based analysis of presence–absence polymorphisms
An iSelect 90 K wheat SNP genotyping array developed by Wang et al. (2014) was used to assess the presence or absence of fluorescent signals from probes known to be specific to chromosomes 7A, 7B or 7D of bread wheat.
RNA qPCR analysis
RNA for quantitative PCR analysis was extracted from wheat flag leaves of plants grown in the field. A Spectrum Plant Total RNA Kit (Sigma-Aldrich) was used for RNA extraction and On-column DNase I Digestion Kit used to remove genomic DNA (Sigma-Aldrich). cDNA was produced by reverse transcription using a Phusion RT-PCR kit (Finnzymes) and qPCR undertaken using a CFX96 Real Time System and C1000 Touch Thermo Cycler (Bio‐Rad). Lr34 transcripts were normalized relative to the wheat glyceraldehyde 3-phosphate dehydrogenase gene (TaGAPDH; GenBank AF251217) using the comparative CT method (Schmittgen and Livak 2008). Primer sequences are shown in Table S1.
DNA blot analysis
DNA from durum seedlings was isolated using the CTAB method (Saghi-Maroof et al. 1984). DNAs were digested with EcoRI, prior to 0.8% agarose gel electrophoresis and then transferred to nylon membranes and hybridized as described by Collins et al. (1998). The Lr34 DNA probe was PCR amplified using primers listed in Table S1 to produce a 372 bp product that encompassed nucleotides 64,619–64,240 of the Lr34 genomic sequence (GenBank FJ436983.1). The probe was labeled with [P32]-dCTP using a Megaprime DNA labelling system (Amersham).
Cytogenetic analysis
Fluorescence in situ hybridisation (FISH) and genomic in situ hybridisation (GISH) analyses were undertaken on plant root tip cells as previously described (Zhang et al. 2001, 2004; Lang et al. 2018). A, B and D genome chromosomes were distinguished in GISH analysis using fluorescein-12dUTP (fluorescent green colour) (Roche Diagnostics Gmbh, Mannheim, Germany) and tetramethyl-rhodamine-5-dUTP (red colour) labelled probes derived from total genomic DNA of T. urartu and Aegilops tauschii, respectively. Individual chromosomes were identified by FISH analysis with oligonucleotide probes, oligo-pSc119.2-1 (green) and oligo-pTa535-1 (red) (Tang et al. 2014).
Results
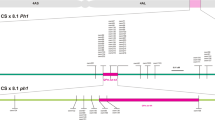
Tetraploid wheat chromosome substitution line LDN 7D(7A) was crossed to Cappelli ph1c (Riley and Chapman 1958; Giorgi and Barberra 1981), and a crossing and screening strategy similar to that reported by Han et al. (2016) was undertaken (Fig. 1a). Chromosome 7D present in LDN 7D(7A) was derived from the hexaploid wheat cultivar Chinese Spring which encodes the Lr34 gene (Joppa and Williams 1988). F1 plants were backcrossed to the Cappelli ph1c mutant and BC1 plants identified that were, firstly homozygous for ph1c as evidenced by the absence of a wild type Ph1 PCR product (Wang et al. 2002) and secondly, contained chromosome 7D as indicated by an Lr34 marker derived from the cloned Lr34 gene sequence (Table S1) (Fig. 1b) (Lagudah et al. 2009). Progenies (BC1F2) were generated from two BC1 plants by self-fertilisation with 101 progeny from the first BC1 plant and 81 progeny from the second BC1 plant subsequently analysed.
a Crossing strategy used for introgression of the bread wheat chromosome 7D region that encodes Lr34 gene into chromosome 7A of durum wheat. b Molecular marker analysis of BC1 plants produced by crossing LDN 7D(7A) plants with Cappelli ph1c and using Cappelli ph1c as a recurrent parent. The upper panel depicts PCR products obtained with a dominant Lr34 PCR marker derived from the cloned Lr34 gene sequence and amplified using primers ABCTF4N (Krattinger et al. 2009) and Lr34 PLUSR (Lagudah et al. 2009), while the lower panel shows gel electrophoresis of products obtained for a dominant PCR marker specific for the wild type Ph1 gene. This latter marker also amplified nonspecific products in the Cappelli ph1c deletion mutant. Each panel shows amplification products from LDN 7D(7A) (lane 2), Cappelli ph1c (lane 3) and BC1 plants (lanes 4–6). A 1 kb Plus molecular weight DNA marker (M) (Invitrogen) is shown in lane 1
As a preliminary screen DNAs from BC1F2 plants were subject to real time qPCR whereby a wheat D genome-specific repeat sequence, Dgas44 (McNeil et al. 1994) was quantified relative to the wheat 28S rRNA genes (Han et al. 2016) (Fig. 2a, b). These plant DNAs were also screened for the presence of Lr34 using the marker described above (Fig. 1b; Table S1). From this analysis, 15 and 16 Lr34 positive plants were identified from each cross (14.8% and 19.7% of plants screened, respectively) that contained 2–10% of Dgas44 sequences compared with the homozygous LDN 7D(7A) parent substitution line (Fig. 2c, d). Potentially these plants contained small 7D introgressions in chromosome 7A that encode Lr34. Given the relatively large number of potential recombinants identified in these two families only progeny from the first family were analysed further as these plants were planted earlier and hence more mature.
a Quantification of a D genome-specific repeat sequence, Dgas44, in LDN 7D(7A) x Cappelli ph1c BC1F2 progeny from BC1 plant 1 using qPCR. Each column depicts the relative amount of Dgas44 amplification product obtained from a single progeny plant. Dgas44 amplification products were normalised relative to a control wheat gene (GAPDH) and this ratio was then normalised relative to the Dgas44/GAPDH value obtained for LDN 7D(7A). The LDN 7D(7A) value (100%) is shown as a vertical black column that is arrowed on the right whereas the value obtained for a LDN control line is highlighted by an arrow on the left. Red lines indicate plants containing Lr34 and blue lines show plants without Lr34. b Graph depicting the same Dgas44 assay shown in (A) undertaken on progeny from BC1 plant 2. c Graph of the same plants shown in (A) that contain Lr34 and low amounts of Dgas sequence. d Graph of the same plants shown in (B) containing Lr34 and low amounts of Dgas44 sequence (colour figure online)
To confirm the results obtained from Dgas44 qPCR a series of dominant and co-dominant KASP markers were used to screen 71 BC1F2 plants from this family. These markers confirmed that frequent homoeologous recombination events occurred between chromosomes 7A and 7D in the ph1c mutant background (Fig. S1). From the analysis of these 71 plants, 46 (65%) contained at least one recombinant 7D/7A chromosome (Fig. S1). Of the 15 plants originally identified as Lr34 positive and containing less than 10% Dgas44 (Fig. 1c), 11 underwent this KASP analysis and all were confirmed to have small 7D introgressions (Fig. S1, marked with a black asterisk).
A set of 16 Lr34 positive plants were selected for further analysis. These included 11 of the 15 plants that had Dgas44 levels below 10% (Fig. 2c) and for which supporting KASP marker data was successfully obtained (Fig. S1, black asterisk). Plant 43-2 was included, which contained 13% Dgas44 and only three of 16 7D KASP alleles (Fig. S1, marked with X). Finally, four plants (97-4, 97-5, 97-17, 133-27) were also further analysed as they potentially contained a chromosome with a small Lr34 introgression, in addition to a complete 7D chromosome (Fig. S1, red asterisks).
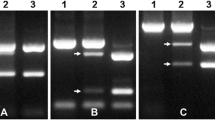
Four 7DS PCR markers that resolved LDN 7D(7A) from both LDN and Cappelli ph1c were identified as described in the materials and methods. Three of these markers (Cs7D1, Cs7D2, Cs7D4) were codominant and one dominant (Cs7D3) (Fig. 3a). These markers were screened on the 16 plants described above and the PCR and KASP marker data combined for genotyping. Four plants (97-33, 133-21, 133-25 and 133-28) had similar sized introgressions carrying Lr34 and contained less than 8% of chromosome 7D based upon physical marker positions and the CS reference genome sequence v1.0. Of these plants, one (133-28) was homozygous for three co-dominant 7D markers (wri855, Cs7D1 and Cs7D2) indicating it had inherited two recombinant chromosomes, although the zygosity for Lr34 could not be established given the dominant nature of this marker. Two of these five plants (133-21 and 133-25) also contained a 7D marker at the terminus of the long arm of 7A indicating they contained either two recombinant chromosomes or one with two introgressions.
Mapping of chromosome 7D introgressions on chromosome 7A. KASP and PCR markers used to define 7D introgressions in BC1F2 plants are indicated on the left-hand side with nucleotide co-ordinates of each sequence in the CS genome v1.0 shown in parenthesis. Chromosome 7A from plants (labelled at top) are shown diagrammatically (not to scale) with homozygous 7A sequences white, heterozygous 7D/7A sequences grey and homozygous 7D sequences black. The four plants on the right (137-27, 97-5, 97-4 and 97-17) contained a full length 7D chromosome in addition to a recombinant 7D/7A introgression chromosome. The hatched region shown for plants 133-28, 137-27, 97-4 and 97-17 indicate that recombination break points that occurred in this region could not be more accurately determined due the absence of co-dominant 7D/7A markers in the region. Zygosity of dominant markers for the remaining chromosomes was assumed based on the genotypes of surrounding co-dominant markers, a strategy consistent with the least number of independent recombination events having occurred. Dominant markers are prefaced with (+). The arrow at right indicates approximate centromere position
A further eight plants (35-1, 43-2, 97-10, 97-14, 97-24, 133-2, 133-23, 133-51) were heterozygous for similar sized 7D introgressions that encoded less than 31% of chromosome 7D with one plant also containing the distal end of 7DL (Fig. 3, 97-14). Of the four plants (97-4, 97-5, 97-17 and 137-27) that contained a complete chromosome 7D in addition to a 7D/7A recombinant one (Fig. 3) plant 97-5 was likely homozygous for Lr34, whereas the recombination break point and Lr34 zygosity could not be determined for the other three plants due to a lack of co-dominant markers in the region (Fig. 3).
Four heterozygous BC1F2 plants that carried Lr34 (97-14, 97-33, 133-21, 133-25) were self-fertilised and homozygous BC1F3 seedlings selected using codominant markers Cs7D1, Cs7D2 and Cs7D4 (Fig. 3; Table S1). DNAs from these four homozygous progenies (97-14-9, 97-33-1, 133-21-2, 133-25-9) were then assayed using wheat Infinium iSelect 90 K SNP arrays to further resolve both the size of introgressed 7D segments and the number of introgressions within the chromosome. SNP markers were identified that were polymorphic between the LDN and Cappelli ph1c genomes and also a set that was specific to the LDN 7D(7A) chromosome substitution line (i.e., 7D specific) (Fig. 4).
90 K SNP marker analysis of 7D and 7A chromosomes in selected homozygous BC1F3 plants (97-14-9, 97-33-1, 133-21-2, 133-25-9). Markers are shown as grey lines, or boxes when groups of markers are present, on each chromosome with CS v1.0 genome nucleotide co-ordinates shown as arrowheads on the left of each chromosome pair with each arrowhead depicting 100 Mb intervals. Karyotypes 1–6 are as follows; (1) 7D specific markers present on chromosome 7D of the parental LDN 7D(7A) substitution line (all 7D), (2) 7D markers present in the BC1F3 plants indicated (7D present), (3) 7A markers present in the BC1F3 plants (7A present), (4) 7A markers absent in BC1F3 plants (7A absent), (5) 7B markers present in BC1F3 plants (7B present), (6) 7B markers absent in the BC1F3 plants (7B absent)
Chromosome 7D specific markers were identified in these plant DNAs that further confirmed the presence of 7D introgressions (Fig. 4) and were generally consistent with the initial marker mapping results (compare Figs. 3 and 4). Evidence for homozygosity and the presence of 7DS introgessions in plants 97-33-1, 133-21-2 and 133-25-9 was supported by the absence of 7A markers located in introgressed 7DS regions and presence of 7A markers on the remainder of the chromosome (Fig. 4). This SNP marker pattern showed slightly less correlation for plant 97-14-9 for unknown, presumably technical reasons. No evidence of introgressions into chromosome 7B for any of the plants analysed was observed (Fig. 4). Plant 133-21-2 was homozygous for a double recombinant chromosome with 7D introgressions present at both ends of 7A, given the absence of 7A markers in the corresponding regions (Fig. 4). Finally, 90 K SNP analysis (Fig. 4) showed that plants 97-14-9 and 133-25-9 were both homozygous for a recombinant chromosome containing only a single 7DS integration suggesting their parents were likely to contain two separate recombinant chromosomes rather than a single recombinant chromosome with two 7D introgressions (Fig. 4).
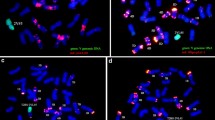
FISH and GISH analyses were then undertaken on BC1F3 plants 97-14-3 and 97-33-1 which are homozygous for 7DS introgressions derived from plants 97-14 and 97-33, respectively. Upon hybridisation with Ae. tauschii (DD) genomic DNA as a probe, small chromosome 7D introgressions were clearly identified (Fig. 5a). Subsequent FISH analysis demonstrated that these 7D introgressions were in chromosome 7A in each case. Therefore, plants 97-14-3 and 97-33-1 were each homozygous for a single distal 7DS introgression in chromosome 7A, consistent with molecular marker analysis undertaken on sibs of these plants described above.
Cytogenetic and DNA blot analyses of Lr34 durum wheat plants. a Cytological analysis of root tip chromosome spreads from two BC1F3 plants, 97-14-3 (upper panels) and 97-33-1 (lower panels). GISH chromosomes on the left were hybridised with A and D genome specific probes generating green and red signals, respectively, whereas FISH images on the right were from hybridisation with oligo-pTA535-1 (red label) and pSc119.2-1 (green label) to identify individual chromosomes. Arrow heads indicate introgressed chromosome 7D segments that show red fluorescence in GISH images (1 and 3). b, c Segregation of the Lr34 gene in progeny of Bansi Lr34-13 and Bansi Lr34-15 plants. DNAs were digested with EcoRI and hybridised with a Lr34 probe. Arrow heads at the left of each figure indicate the chromosome origin of each restriction fragment based upon the CS genome sequence v1.0. Note that the 7B homoeologue of Lr34 is thought to have translocated to chromosome 4A (Krattinger et al. 2011). In panel (B), lanes 1–7 contain DNAs from progeny of Bansi plant Lr34-13 while lanes 8–10 contain genomic DNAs of bread wheat cv. Fielder, durum chromosome substitution line LDN 7D(7A) and bread wheat cv. Robin, respectively. In panel (C) lanes 1–12 contain DNAs from progeny of line Bansi Lr34-15, whereas lanes 13–17 contain genomic DNAs from chromosome substitution line LDN 7D(7A), wild type LDN, Cappelli ph1c durum, durum cv. Bansi and bread wheat cv. Robin, respectively. Note a presence/absence polymorphism for the 7A Lr34 RFLP fragment exists between durum lines Cappelli ph1c, LDN and Bansi which is described further in Fig. S3. In panels (B) and (C) lanes 11 and 18, respectively, show molecular weights of 6, 5, 4, 3 and 2 kb. The red line in panel (B) indicates the point at which additional lanes on the same autoradiograph were removed as they were not relevant to the final figure. d Molecular characterisation of Lr34 gene sequences amplified from genomic DNA of Bansi Lr34-13-1. Coding sequences of the Lr34 gene were amplified as five separate fragments (blue lines) using the primer sequences indicated at the end of each amplification product. Each amplification product was sequenced using primers depicted as black arrows with sequence direction shown as hatched black lines. All exons were sequenced in their entirety. Primer sequences and their full names are shown in Table S1, while abbreviated names for primers are used in this figure (colour figure online)
Plant 97-14-3 was crossed using Bansi as a recurrent parent for 3 generations and segregation of the 7D/7A Lr34 gene was apparent in progeny derived from selfed BC2 Bansi plants, Bansi Lr34-13 and Bansi Lr34-15, by DNA blot (Fig. 5b, c). Two plants homozygous for the Lr34 D chromosome introgression in this generation, Bansi Lr34-13-1 and Bansi Lr34-15-9, and a null sib (Bansi Lr34-13-7 null) were selected for progeny field testing in 2021. Visual scoring showed that the progeny of Bansi Lr34-13-1 and Bansi Lr34-15-9 were susceptible as was the Bansi parent (Fig. 6a). These visual scores were confirmed on the same field material using a chitin-based fungal biomass assay (Fig. 6a). The Lr34 genotypes of this material was also reconfirmed by using Lr34 specific markers on tissue taken from the field plots (Fig. S2).
Field trial and expression analysis of Lr34 durum plants. a The graph at left shows fungal biomass quantification of flag leaves from field grown (2021) durum and bread wheat plants. The Y-axis shows fluorescence units with increasing fluorescence equal to increasing fungal biomass. The numbers at the top of the graph indicate visual scores (VS) assigned to each genotype for stripe rust response, on a 0 (immune) to 9 (fully susceptible) scale. The same tissue was used for both biomass quantification and stripe rust response assessment. RNA was also extracted from flag leaves of the same material and Lr34 expression determined by qRT-PCR (graph on the right). Common letters above columns indicate no significant difference between values (ANOVA with posthoc Tukey). b Stripe rust response scores (left hand axis, black columns) and fungal biomass quantification (right hand axis, grey columns) of wheat genotypes grown in 2022. The same field grown material was used for both tests. Common letters above columns indicate no significant difference between values (ANOVA with posthoc Tukey test, p < 0.1). Note that statistical comparisons were made only between plants within each of the following groups: [Tc, Tc34], [LDN, Cappelli ph1c, durum 97 plants] and [Svevo, Svevo BC plants]. These three groups were not intercompared given their different genetic backgrounds. Biomass data and visual pathology data were compared separately. c Quantitative RT-PCR analysis of Lr34 expression in wheat plants grown in the 2022 field season. Expression levels are relative to the wheat GAPDH gene. Expression analysis was undertaken on tissue isolated from the same plants used in (B) above. Samples with common letters above each column are not significantly different (ANOVA, post-hoc Tukey tests)
The Lr34 gene carried by LDN 7D(7A) was originally derived from bread wheat cultivar Chinese Spring (Joppa and Williams 1988). To ensure an equivalent, functional version of the gene was present in the Bansi durum material the Lr34 ORF was PCR amplified and sequenced from genomic DNA of line Bansi Lr34-13-1. The coding regions of the gene were amplified from this durum line as five discrete fragments and sequenced (Fig. 5d). All exon sequences were identical to the published Chinese Spring sequence (GenBank FJ436983.1) as were the intervening intron sequences present in these fragments.
These Bansi families were retested in 2022 along with homozygous families carrying introgressions derived from two Cappelli/LDN 7D(7A) plants, 97-14 and 97-33, and two Svevo Lr34 lines, Svevo Lr34-32 and Svevo Lr34-46. Svevo Lr34 lines were produced by crossing plant 97-33-1 to Svevo and backcrossing for 6 generations, followed by the selection of homozygous plants from a selfed family of a BC6 plant. These Svevo Lr34 lines contain a smaller 7D introgression derived from plant 97-33, compared with Bansi Lr34 lines and other 97-14 derived material (Fig. 4). Visual scoring and chitin biomass assays again showed no evidence of resistance in Bansi Lr34 material nor was resistance apparent in progeny of Lr34 carrying Cappelli/LDN 7D(7A) plants 97-14-3-13 and 97-33-1-9 compared with the parental controls (Fig. 6b). In contrast, Svevo Lr34 and control Svevo lines showed high levels of resistance (Fig. 6b) indicating that Svevo contains pre-existing resistance to these Pst isolates that masked any potential resistance conferred by the 7DS segment. Accelerated leaf tip necrosis was not observed in Lr34 durum lines compared with controls.
The absence of Lr34 resistance in these durum materials was further investigated by expression analysis. RNA was extracted from flag leaves of the infected field materials grown in 2021 and quantitative RT-PCR undertaken using primers specific for the Lr34 resistance allele. An obvious, four–fivefold reduction in Lr34 expression was observed in Bansi Lr34 durum plants when compared with hexaploid bread wheat Thatcher Lr34 plants, grown under the same field conditions (Fig. 6a). An RNA expression analysis was also undertaken using leaf material from the 2022 field trial and again Thatcher Lr34 lines showed significantly higher expression of Lr34 compared with Bansi or Cappelli/LDN 7D(7A) derivatives containing this APR gene (Fig. 6c). These data demonstrate a significantly lower level of Lr34 expression in these durum plants in the field when compared with the endogenous gene in bread wheat.
Discussion
There are limited examples of transfer of genes from the D genome of T. aestivum to T. turgidum. These examples include the introduction of seed storage protein genes for improved dough quality (Lukaszewski and Curtis 1994; Joppa et al. 1998; Ceoloni et al. 1996; Liu et al. 1996; Lukaszewski 2003), the KNa1 gene for improved Na+ tolerance (Dvorak and Gorham 1992; Luo et al. 1996), the TaAlmt1 gene for improved Al3+ tolerance (Han et al. 2014, 2016) and random D-genome fragments (Eberhard et al. 2010; Othmeni et al. 2019). These studies used a mixture of cytogenetics, protein markers, RFLP markers, SNP markers, PCR markers, DArT markers or Dgas44 qPCR, for identifying and defining introgressions.
Here we used three approaches to identify and define chromosome 7D introgressions into chromosome 7A. Each approach had specific limitations in that KASP and PCR marker analysis could not identify all 7D introgressions due to the limited marker coverage available at the time, 90 K SNP analysis required extensive bioinformatic analysis and cytogenetic analysis resulted in coarse breakpoint refinement. None the less, a combination of these approaches unambiguously identified small, single 7D introgressions in chromosome 7A that encode Lr34, with the introgression present in plant 97-33-1 estimated to account for less than 8% of chromosome 7D.
Field trials showed no enhanced adult plant resistance to Pst in Cappelli/LDN 7D(7A) or Bansi durum plants carrying Lr34 on the introgressed 7D/7A segment. Lr34 expression in flag leaves of field grown Cappelli/LDN 7D(7A) and Bansi durum plants was four–fivefold lower than that observed in hexaploid Thatcher-Lr34 plants grown in the same field plots. Presumably this low level of expression accounted for the inability of Lr34 to function in these durum plants and to provide stripe rust resistance. These Lr34 carrying durum lines have not been challenged with either leaf rust, stem rust, or powdery mildew. Therefore, their potential resistance to these other diseases remains unknown.
There is an established correlation between Lr34 expression levels and resistance. For example, increased Lr34 expression occurs in adult plant tissues of bread wheat when compared with seedlings, which presumably accounts for the resistance being observed in mature tissues, only (Krattinger et al. 2009). In addition, bread wheat flag leaf tips show higher expression than do flag leaf bases which also corelates with increased disease susceptibility of the latter tissue (Krattinger et al. 2009). Similarly, Lr34 resistance to P. triticina can be observed in bread wheat seedlings when grown at low temperature (10 °C), which also correlates with higher levels of Lr34 expression (Rinaldo et al. 2017). As already noted, Lr34 mediated resistance in transgenic durum wheat was also strongly correlated with transgene expression levels (Rinaldo et al. 2017) as were Lr34 pleotropic phenotypes in heterologous host species (Risk et al. 2013; Krattinger et al. 2016; Schnippenkoetter et al. 2017; Sucher et al. 2017).
Suppression of wheat disease resistance genes in new genetic backgrounds is not uncommon, particularly when resistance genes are transferred from diploid and tetraploid species to bread wheat (Hiebert et al. 2020 and references therein). Recently, one example was shown to be due to a D genome gene, Med15, which encodes a subunit of the transcriptional coactivation mediator complex, that altered gene transcription and suppressed plant immunity (Hiebert et al. 2020). Genetic background was also shown to affect the efficacy of the Fhb1 Fusarium head blight resistance gene when introgressed into durum wheat from bread wheat (Kirana et al. 2023), while the rye Pm8 powdery mildew resistance gene is suppressed by the orthologous wheat Pm3 gene via a post-translational mechanism (McIntosh et al. 2011; Hurni et al. 2014). Recent genomic and transcriptomic analyses have also shown that genes present on introgressed segments in bread wheat can have highly variable expression patterns. In bread wheat plants containing chromosome segments from Ambylopyrum muticum, less than half the introgressed genes were expressed (Coombes et al. 2022). Introgressions from Ae. longissima were shown to change expression of approximately 4% of endogenous wheat genes located elsewhere in the genome (Dong et al. 2020). Similarly in rice, changes in DNA methylation and gene expression were associated with homoeolog exchange in allotetraploid plants (Li et al. 2019). The interplay between endogenous and homoeologous plant chromatin segments is therefore complex.
The basis for the suppressed Lr34 expression observed here for two independent 7D/7AS introgressions in durum wheat is unknown. Clearly useful genes present in alien introgressions do not always show suppressed expression. Since E.R. Sears (1956), pioneered interspecies introgressions into wheat there have been numerous introgressions into durum (see above) and bread wheat that provide useful agronomic traits, particularly for disease resistance (Rather et al. 2017; Baranwal 2022). In fact, “linkage drag” caused by expression of additional unwanted genes present on introgressed segments can often be problematic. However, in this study, the Lr34 APR trait of interest is not manifest in the durum wheat backgrounds examined due to suppressed gene expression.
Data availability
All data and materials are available upon request.
References
Ayliffe M, Singh R, Lagudah E (2008) Durable resistance to stem rust needed. Curr Opin Plant Biol 11:187–192
Ayliffe M, Periyannan SK, Feechan A, Dry I, Schumann U, Wang M-B, Pryor A, Lagudah E (2013) A simple method for comparing fungal biomass in infected plant tissues. Mol Plant Microbe Inter 26:658–667
Ayliffe M, Luo M, Faris J, Lagudah E (2022) Disease Resistance. In: Reynolds, M.P., Braun, HJ. (eds) Wheat Improvement. Springer, Cham. https://doi.org/10.1007/978-3-030-90673-3_19
Baranwal D (2022) Genetic and genomic approaches for breeding rust resistance in wheat. Euphytica 218:159
Bariana HS, Miah H, Brown GN, Willey N, Lehmensiek A (2007) Molecular mapping of durable rust resistance in wheat and its implication in breeding. In: Buck HT, Nisi JE (eds) Wheat production in stressed environments. Developments in Plant Breeding, vol 12. Springer, Dordrecht, pp 723–728
Bhavani S, Hodson DP, Huerta-Espino J, Randhawa MS, Singh RP (2019) Progress in breeding for resistance to Ug99 and other races of the stem rust fungus in CIMMYT wheat germplasm. Front Agric Sci Eng 6:210–224
Bryan GJ, Dixon A, Gale MD, Wiseman G (1998) A PCR-based method for the detection of hexaploid bread wheat adulteration of durum wheat and pasta. J Cereal Sci 28:135–145
Ceoloni C, Biagetti M, Ciaffi M, Forte P, Pasquini M (1996) Wheat chromosome engineering at the 4× level: the potential of different alien gene transfers into durum wheat. Euphytica 89:87–97
Collins N, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor AJ (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:968–978
Coombes B, Fellers JP, Grewai S, Rusholme-Picher R, Hubbart-Edwards S, Yang C-Y, Joynson R, King IP, King J, Hall A (2022) Whole genome sequencing uncovers the structural and transcriptomic landscape of hexaploid/Ambylopyrum muticum introgression lines. Plant Biotech J. https://doi.org/10.1111/pbi.13859
Dong Z, Ma C, Tian X, Zhu C, Wang LY, Friebe B, Li H, Liu W (2020) Genome-wide impacts of alien chromosome introgressions on wheat gene transcriptions. Sci Rep 10:4801
Dvorak J, Gorham J (1992) Methodology of gene transfer by homoelogous recombination into Triticum turgidum: transfer of K+/Na+ discrimination from Triticum aestivum. Genome 35:639–646
Dyck PL, Samborsk DJ, Anderson RG (1966) Inheritance of adult-plant leaf rust resistance derived from common wheat varieties exchange and Frontana. Can J Genet Cytol 8:665–671
Eberhard FS, Zhang P, Lehmensiek A, Hare RA, Simpendorfer S, Sutherland MW (2010) Chromosome composition of an F2 Triticum aestivum X T. turgidum spp. durum cross analysed by DArT markers and MCFISH. Crop Pasture Sci 61:619–624
Giorgi B, Barberra F (1981) Increase of homoeologous pairing in hybrids between a ph mutant of Trictium turgidum L. var durum and two tetraploid species of Aegilops: Aegilops kotschyi and Ae. cylindrica. Cereal Res Commun 9:205–211
Han C, Ryan PR, Yan Z, Delhaize E (2014) Introgression of a 4D chromosomal fragment into durum wheat confers aluminium tolerance. Ann Bot 114:135–144
Han C, Zhang P, Ryan PR, Rathjen TM, Yan Z, Delhaize E (2016) Introgression of genes from bread wheat enhances the aluminium tolerance of durum wheat. Theor Appl Genet 129:729–739
Herrera-Foessel SA, Singh RP, Lillemo M, Huerta-Espino J, Bhavani S, Singh S, Lan C, Calvo-Salazar V, Lagudah ES (2014) Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor Appl Genet 127:781–789
Hiebert CW, Moscou MJ, Hewitt T, Steuernagel B, Hernández-Pinzón I, Green P, Pujol V, Zhang P, Rouse MN, Jin Y, McIntosh RA, Upadhyaya N, Zhang J, Bhavani S, Vrána J, Karafiátová M, Huang L, Fetch T, Doležel J, Wulff BBH, Lagudah E, Spielmeyer W (2020) Stem rust resistance in wheat is suppressed by a subunit of the mediator complex. Nat Commun 11:1123
Huerta-Espino J, Singh RP, German S, McCallum BD, Park RF, Chen WQ, Bhardwaj SC, Goyeau H (2011) Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 179:143–160
Hurni S, Brunner S, Stirnweis D, Herren G, Peditto D, McIntosh RA, Keller B (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J 79:904–913
Joppa LR, Klindworth DL, Hareland GA (1998) Transfer of high molecular weight glutenins from spring wheat to durum wheat, pp. 257–260. In: Proceedings of the 9th international wheat genetics symposium, Vol. 1, Saskatoon, Saskatchewan, Canada, edited by A. E. Slinkard. University of Saskatchewan Extension Press, Saskatoon, SK, Canada
Joppa LR, Williams ND (1988) Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30:222–228
Kirana RP, Michel S, Moreno-AMores J, Prat N, Lemmens M, Buerstmayr M, Buerstmayr H, Steiner B (2023) Pyramiding fusarium head blight resistance QTL from T. aestivum, T. dicoccum and T. dicoccoides in durum wheat. Theor Appl Genet 136:201
Kolmer JA (2005) Tracking wheat rust on a continental scale. Curr Opin Plant Biol 8:441–449
Kolmer JA (2015) First report of a wheat leaf rust (Puccinia triticina) phenotype with high virulence to durum wheat in the Great Plains region of the United States. Plant Dis 99:156
Kolmer JA, Singh RP, Garvin DF, Viccars L, William HM, Huerta-Espino J, Ogbonnaya FC, Raman H, Orford S, Bariana HS, Lagudah ES (2008) Analysis of the Lr34⁄Yr18 rust resistance region in wheat germplasm. Crop Sci 48:1841–1852
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Krattinger SG, Lagudah ES, Wicker T, Risk JM, Ashton AR, Selter LL, Matsumoto T, Keller B (2011) Lr34 multi-pathogen resistance ABC transporter: molecular analysis of homoeologous and orthologous genes in wheat and other grass species. Plant J 65:392–403
Krattinger SG, Sucher J, Selter LL, Chauhan H, Zhou B, Tang M, Upadhyaya NM, Mieulet D, Guiderdoni E, Weidenbach D, Schaffrath U, Lagudah ES, Keller B (2016) The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol J 14:1261
Krattinger SG, Kang J, Braunlich S, Boni R, Chauhan H, Selter LL, Robinson MD, Schmid MW, Wiederhold E, Hensel G, Kumlehn J, Sucher J, Martinoia E, Keller B (2019) Abscisic acid is a substrate of the ABC transporter encoded by the durable wheat disease resistance gene Lr34. New Phytol 223:853–866
Lagudah ES, Krattinger ES, Spielmeyer W, Singh RP, Huerta-Espino J, Spielmeyer W, Brown-Guiedira G, Selter LL, Keller B (2009) Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor Appl Genet 119:889–898
Lang T, La S, Li B, Yu Z, Chen Q, Li J et al (2018) Precise identification of wheat-Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 61:177–185
Li N, Xu C, Zhang A, Lv R, Meng X, Lin X, Gong L, WendelLiu JFB (2019) DNA methylation repatterning accompanying hybridisation, whole genome doubling and homoeolog exchange in nascent segmental rice allotetraploids. New Phytol 233:979–992
Liu C-Y, Shepherd KW, Rathjen AJ (1996) Improvement of durum wheat pastamaking and breadmaking qualitites. Cereal Chem 73:155–166
Lukaszewski AJ (2003) Registration of six germplasms of durum wheat with introgressions of the Glu-D1 locus. Crop Sci 43:1138–1139
Lukaszewski AJ, Curtis CA (1994) Transfer of the Glu-D1 gene from chromosome 1D to chromosome 1A in hexaploid triticale. Plant Breed 112:177–182
Luo M-C, Dubcovsky J, Goyal S, Dvořák J (1996) Engineering of interstitial foreign chromosome segments containing the K+/Na+ selectivity gene Kna1 by sequential homoeologous recombination in durum wheat. Theor Appl Genet 93:1180–1184
McIntosh R, Zhang P, Cowger C, Park R, Lagudah E, Hoxha S (2011) Rye-derived powdery mildew resistance gene Pm8 in wheat is suppressed by the Pm3 locus. Theor Appl Genet 123:359–367
McNeil D, Lagudah ES, Hohmann U, Appels R (1994) Amplification of DNA-sequences in wheat and its relatives: the Dgas44 and R350 families of repetitive sequences. Genome 37:320–327
Olivera P, Newcomb M, Szabo LJ, Rouse M, Johnson J, Gale S, Luster DG, Hodson D, Cox JA, Burgin L, Hort M, Gilligan CA, Patpour M, Justesen AF, Hovmøller MS, Woldeab G, Hailu E, Hundie B, Tadesse K, Pumphrey M, Singh RP, Jin Y (2015) Phenotypic and genotypic characterisation of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013–4. Phytopathology 105:917–928
Othmeni M, Grewal S, Hubbart-Edwards S, Yang C, Scholefield D, Ashling S, Yahyaoui A, Gustafson P, Singh PK, King IP, King J (2019) The use of pentaploid crosses for the introgression of the Amblyopyrum muticum and D-genome chromosome segments into durum wheat. Front Plant Sci 10:1110
Patpour M, Hovmoller MS, Hodson D (2017) First report of virulence to Sr25 in race TKTTF of Puccinia graminis f. sp. tritici causing stem rust on wheat. Plant Dis 101:1678
Rasheed A, Wen W, Gao F, Zhai S, Jin H, Liu J, Guo Q, Zhang Y, Dreisigacker S, Xia X, He Z (2016) Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor Appl Genet 129:1843–1860
Rather SA, Sharam D, Joshi N (2017) Alien gene introgression in wheat, pp 101–120 Publisher: Kalyani Publishers, New Delhi-110002
Riley R, Chapman V (1958) Genetic control of the cytological diploid behavior of hexaploid wheat. Nature 182:713–715
Rinaldo A, Gilbert B, Boni R, Krattinger S, Singh D, Park R, Lagudah E, Ayliffe M (2017) The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol J 15:894–905
Risk JM, Selter LL, Chauhan H, Krattinger SG, Kumlehn J, Hensel G, Viccars LA, Richardson TM, Buesing G, Troller A, Lagudah ES, Keller B (2013) The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol J 11:847–854
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard R (1984) Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Schnippenkoetter W, Lo C, Liu G, Dibley K, Chan WL, White J, Milne R, Zwart A, Kwong E, Keller B, Godwin I, Krattinger SG, Lagudah E (2017) The wheat Lr34 multipathogen resistance gene confers resistance to anthracnose and rust in sorghum. Plant Biotechol J 15:1387–1396
Sears ER (1977) An induced mutant with homoeologous pairing in common wheat. Can J Genet Cytol 19:585–593
Sears ER (1956) The transfer of leaf rust resistance from Aegilops umbellulata to wheat. Genetics in plant breeding. Brook-haven symposia in biology, pp 1–22
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481
Singh RP, Hodson DP, Jin Y, Lagudah ES, Ayliffe MA, Bhavani S, Rouse MN, Pretorius Z, Huerta-Espino J, Basnet BR, Lan C (2015) Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105:844–872
Sucher J, Roni R, Yang P, Rogowsky P, Buchner H, Kastner C, Kumlehn J, Krattinger SG, Keller B (2017) The durable wheat disease resistance gene Lr34 confers common rust and northern corn leaf blight resistance in maize. Plant Biotechnol J 15:489–496
Tang Z, Yang Z, Fu S (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119, pTa-535, pTa71, CCS1, and pAWRC for FISH analysis. J Appl Genet 55:313–318
Wang XW, Lai JR, Lui GT, Chen F (2002) Development of a SCAR marker for the Ph1 locus in common wheat and its application. Crop Sci 42:1365–1368
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S et al (2014) Characterisation of polyploid wheat genomic DNA diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Watkins JL, Li M, McQuinn RP, Chan KX, McFarlane HE, Ermakova M, Furbank RT, Mares D, Dong C, Chalmers KJ, Sharp P, Mather DE, Pogson BJ (2019) A GDSL esterase/lipase catalyzes the esterification of lutein in bread wheat. Plant Cell 31:3092–3112
Zhang P, Friebe B, Lukaszewski AJ, Gill BS (2001) The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110:335–344
Zhang P, Li W, Friede B, Gill BS (2004) Simultaneous painting of three genomes in hexaploid wheat by BAC-FISH. Genome 47:979–987
Acknowledgements
We wish to thank the China Scholarship Council for providing a PhD scholarship to HL.
Funding
Open access funding provided by CSIRO Library Services. This research was funded by CSIRO and the China Scholarship Council.
Author information
Authors and Affiliations
Contributions
HL, BB and MA produced families and mapped introgressions. DM, AB mapped KASP markers, PZ, JL, SS undertook cytology and field pathology tests, SH undertook backcrossing, SC produced DNA blots, ML undertook qPCR and KF did SNP analysis and mapping. LZ undertook student (HL) supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Steven S. Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, H., Zhang, P., Luo, M. et al. Introgression of the bread wheat D genome encoded Lr34/Yr18/Sr57/Pm38/Ltn1 adult plant resistance gene into Triticum turgidum (durum wheat). Theor Appl Genet 136, 226 (2023). https://doi.org/10.1007/s00122-023-04466-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04466-z