Abstract
Soil salinity is a major threat to crop productivity and quality worldwide. In order to reduce the negative effects of salinity stress, it is important to understand the genetic basis of salinity tolerance. Identifying new salinity tolerance QTL or genes is crucial for breeders to pyramid different tolerance mechanisms to improve crop adaptability to salinity. Being one of the major cereal crops, wheat is known as a salt-sensitive glycophyte and subject to substantial yield losses when grown in the presence of salt. In this study, both pot and tank experiments were conducted to investigate the genotypic variation present in 328 wheat varieties in their salinity tolerance at the vegetative stage. A Genome-Wide Association Studies (GWAS) were carried out to identify QTL conferring salinity tolerance through a mixed linear model. Six, five and eight significant marker-trait associations (MTAs) were identified from pot experiments, tank experiments and average damage scores, respectively. These markers are located on the wheat chromosomes 1B, 2B, 2D, 3A, 4B, and 5A. These tolerance alleles were additive in their effects and, when combined, increased tolerance to salinity. Candidate genes identified in these QTL regions encoded a diverse class of proteins involved in salinity tolerance in plants. A Na+/H+ exchanger and a potassium transporter on chromosome 5A (IWB30519) will be of a potential value for improvement of salt tolerance of wheat cultivars using marker assisted selection programs. Some useful genotypes, which showed consistent tolerance in different trials, can also be effectively used in breeding programs.

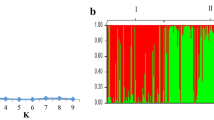
taken from the current experiment)





Similar content being viewed by others
References
Acosta-Motos JR, Ortuño MF, Bernal-Vicente A, Diaz-Vivancos P, Sanchez-Blanco MJ, Hernandez JA (2017) Plant responses to salt stress: adaptive mechanisms. Agron 7:18
Almeida P, Katschnig D, De Boer AH (2013) HKT transporters—state of the art. Int J Mol Sci 14:20359–20385
Alqudah AM, Sallam A, Baenziger PS, Börner A (2020) GWAS: Fast-forwarding gene identification and characterization in temperate Cereals: lessons from Barley–A review. J Adv Res 22:119–135
Ammar M, Pandit A, Singh R, Sameena S, Chauhan M, Singh A, Sharma P, Gaikwad K, Sharma T, Mohapatra T (2009) Mapping of QTLs controlling Na+, K+ and CI− ion concentrations in salt tolerant indica rice variety CSR27. J Plant Biochem Biotechnol 18:139–150
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36:229–239
Bates D, Mächler M, Bolker B, Walker S (2014) Fitting linear mixed-effects models using lme4. arXiv preprint arXiv:14065823
Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1; 1. PLoS Genet 6:e1001193
Borjigin C, Schilling RK, Bose J, Hrmova M, Qiu J, Wege S, Situmorang A, Byrt C, Brien C, Berger B (2020) A single nucleotide substitution in TaHKT1; 5-D controls shoot Na+ accumulation in bread wheat. Plant Cell Environ 43:2158–2171
Brini F, Hanin M, Mezghani I, Berkowitz GA, Masmoudi K (2007) Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt-and drought-stress tolerance in Arabidopsis thaliana plants. J Exp Bot 58:301–308
Bulle M, Yarra R, Abbagani S (2016) Enhanced salinity stress tolerance in transgenic chilli pepper (Capsicum annuum L.) plants overexpressing the wheat antiporter (TaNHX2) gene. Mol Breed 36:36
Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R (2007) HKT1; 5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 143:1918–1928
Byrt CS, Xu B, Krishnan M, Lightfoot DJ, Athman A, Jacobs AK, Watson-Haigh NS, Plett D, Munns R, Tester M (2014) The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J 80:516–526
Chaurasia S, Singh AK, Songachan L, Sharma AD, Bhardwaj R, Singh K (2020) Multi-locus genome-wide association studies reveal novel genomic regions associated with vegetative stage salt tolerance in bread wheat (Triticum aestivum L.). Genomics 112:4608–4621
Chen ZC, Yamaji N, Fujii-Kashino M, Ma JF (2016) A cation-chloride cotransporter gene is required for cell elongation and osmoregulation in rice. Plant Physiol 171:494–507
Chen T, Zhu Y, Chen K, Shen C, Zhao X, Shabala S, Shabala L, Meinke H, Venkataraman G, Chen ZH (2020) Identification of new QTL for salt tolerance from rice variety Pokkali. J Agron Crop Sci 206:202–213
Choudhury S, Larkin P, Xu R, Hayden M, Forrest K, Meinke H, Hu H, Zhou M, Fan Y (2019) Genome wide association study reveals novel QTL for barley yellow dwarf virus resistance in wheat. BMC Genom 20:1–8
Chu W, Li R, Liu J, Reimherr M (2020) Feature selection for generalized varying coefficient mixed-effect models with application to obesity GWAS. Ann Appl Stat 14:276–298
Cuin TA, Betts SA, Chalmandrier R, Shabala S (2008) A root’s ability to retain K+ correlates with salt tolerance in wheat. J Exp Bot 59:2697–2706
Cuin TA, Parsons D, Shabala S (2010) Wheat cultivars can be screened for NaCl salinity tolerance by measuring leaf chlorophyll content and shoot sap potassium. Funct Plant Biol 37:656–664
Dang Y, Routley R, McDonald M, Dalal R, Singh D, Orange D, Mann M (2006) Subsoil constraints in Vertosols: crop water use, nutrient concentration, and grain yields of bread wheat, durum wheat, barley, chickpea, and canola. Aust J Agric Res 57:983–998
Darko E, Gierczik K, Hudak O, Forgó P, Pál M, Türkösi E, Kovács V, Dulai S, Majlath I, Molnar I (2017) Differing metabolic responses to salt stress in wheat-barley addition lines containing different 7H chromosomal fragments. PLoS One 12:e0174170
Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137:807–818
Devi R, Ram S, Rana V, Malik VK, Pande V, Singh GP (2019) QTL mapping for salt tolerance associated traits in wheat (Triticum aestivum L.). Euphytica 215:1–23
Díaz De León JL, Escoppinichi R, Geraldo N, Castellanos T, Mujeeb-Kazi A, Röder MS (2011) Quantitative trait loci associated with salinity tolerance in field grown bread wheat. Euphytica 181:371–383
Dubcovsky J, Santa Maria G, Epstein E, Luo M-C, Dvořák J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet 92:448–454
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fan Y, Shabala S, Ma Y, Xu R, Zhou M (2015) Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genom 16:1–11
Fan Y, Zhou G, Shabala S, Chen Z-H, Cai S, Li C, Zhou M (2016) Genome-wide association study reveals a new qtl for salinity tolerance in barley (Hordeum vulgare L.). Front Plant Sci. https://doi.org/10.3389/fpls.2016.00946
Flowers TJ, Munns R, Colmer TD (2015) Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann Bot 115:419–431
Genc Y, Oldach K, Verbyla AP, Lott G, Hassan M, Tester M, Wallwork H, McDonald GK (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor Appl Genet 121:877–894
Genc Y, Taylor J, Lyons G, Li Y, Cheong J, Appelbee M, Oldach K, Sutton T (2019) Bread wheat with high salinity and sodicity tolerance. Front Plant Sci 10:1280
Ghonaim MM, Mohamed HI, Omran AA (2021) Evaluation of wheat (Triticum aestivum L.) salt stress tolerance using physiological parameters and retrotransposon-based markers. Genet Resour Crop Evol 68:227–242
Gupta B, Huang B (2014) Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genom
Huang J, Xue C, Wang H, Wang L, Schmidt W, Shen R, Lan P (2017) Genes of ACYL CARRIER PROTEIN family show different expression profiles and overexpression of ACYL CARRIER PROTEIN 5 modulates fatty acid composition and enhances salt stress tolerance in Arabidopsis. Front Plant Sci 8:987
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Hura T, Szewczyk-Taranek B, Hura K, Nowak K, Pawłowska B (2017) Physiological responses of Rosa rubiginosa to saline environment. Water, Air, Soil Pollut 228:81
Imamura T, Yasui Y, Koga H, Takagi H, Abe A, Nishizawa K, Mizuno N, Ohki S, Mizukoshi H, Mori M (2020) A novel WD40-repeat protein involved in formation of epidermal bladder cells in the halophyte quinoa. Commun Biol 3:1–14
James RA, Davenport RJ, Munns R (2006) Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol 142:1537–1547
Jia Q, Zheng C, Sun S, Amjad H, Liang K, Lin W (2018) The role of plant cation/proton antiporter gene family in salt tolerance. Biol Plant 62:617–629
Jiang W, Pan R, Buitrago S, Wu C, Abou-Elwafa SF, Xu Y, Zhang W (2021) Conservation and divergence of the TaSOS1 gene family in salt stress response in wheat (Triticum aestivum L.). Physiol Mol Biol Plants 5:1–16
Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20:586–594
Kaler AS, Gillman JD, Beissinger T, Purcell LC (2020) Comparing different statistical models and multiple testing corrections for association mapping in soybean and maize. Front Plant Sci 10:1794
Kang Y, Barry K, Cao F, Zhou M (2020) Genome-wide association mapping for adult resistance to powdery mildew in common wheat. Mol Biol Rep 47:1241–1256
Keisham M, Mukherjee S, Bhatla SC (2018) Mechanisms of sodium transport in plants—progresses and challenges. Int J Mol Sci 19:647
Kong D, Li M, Dong Z, Ji H, Li X (2015) Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep 34:395–410
Kusmec A, Schnable PS (2018) Farm CPU pp efficient large-scale genomewide association studies. Plant Direct 2:e00053
De Leon TB, Linscombe S, Subudhi PK (2017) Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace ‘Pokkali.’ PLoS One 12:e0175361
Liang C, Zhang X, Chi X, Guan X, Li Y, Qin S, bo Shao H (2011) Serine/threonine protein kinase SpkG is a candidate for high salt resistance in the unicellular cyanobacterium Synechocystis sp. PCC 6803. PLoS One 6:e18718
Lu W, Guo C, Li X, Duan W, Ma C, Zhao M, Gu J, Du X, Liu Z, Xiao K (2014) Overexpression of TaNHX3, a vacuolar Na+/H+ antiporter gene in wheat, enhances salt stress tolerance in tobacco by improving related physiological processes. Plant Physiol Biochem 76:17–28
Luo Q, Zheng Q, Hu P, Liu L, Yang G, Li H, Li B, Li Z (2021) Mapping QTL for agronomic traits under two levels of salt stress in a new constructed RIL wheat population. Theor Appl Genet 134:171–189
Ma L, Zhou E, Huo N, Zhou R, Wang G, Jia J (2007) Genetic analysis of salt tolerance in a recombinant inbred population of wheat (Triticum aestivum L.). Euphytica 153:109–117
Ma X, Liang W, Gu P, Huang Z (2016) Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiol Biochem 106:129–140
Ma F, Xu Y, Ma Z, Li L, An D (2018) Genome-wide association and validation of key loci for yield-related traits in wheat founder parent Xiaoyan 6. Mol Breed 38:1–15
Masoudi B, Mardi M, Hervan EM, Bihamta MR, Naghavi MR, Nakhoda B, Amini A (2015) QTL mapping of salt tolerance traits with different effects at the seedling stage of bread wheat. Plant Mol Biol Rep 33:1790–1803
McDonald GK, Taylor J, Verbyla A, Kuchel H (2013) Assessing the importance of subsoil constraints to yield of wheat and its implications for yield improvement. Crop Pasture Sci 63:1043–1065
Monfared HH, Chew JK, Azizi P, Xue G-P, Ee S-F, Kadkhodaei S, Hedayati P, Ismail I, Zainal Z (2020) Overexpression of a rice monosaccharide transporter gene (OsMST6) confers enhanced tolerance to drought and salinity stress in Arabidopsis thaliana. Plant Mol Biol Rep 38:151–164
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30:360–364
Mushke R, Yarra R, Kirti P (2019) Improved salinity tolerance and growth performance in transgenic sunflower plants via ectopic expression of a wheat antiporter gene (TaNHX2). Mol Biol Rep 46:5941–5953
Naeem M, Iqbal M, Shakeel A, Ul-Allah S, Hussain M, Rehman A, Zafar ZU, Ashraf M (2020) Genetic basis of ion exclusion in salinity stressed wheat: implications in improving crop yield. Plant Growth Regul 92:479–496
Nieves-Cordones M, Alemán F, Martínez V, Rubio F (2010) The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol Plant 3:326–333
Nieves-Cordones M, Alemán F, Martínez V, Rubio F (2014) K+ uptake in plant roots. the systems involved, their regulation and parallels in other organisms. J Plant Physiol 171:688–695
Orton TG, Mallawaarachchi T, Pringle MJ, Menzies NW, Dalal RC, Kopittke PM, Searle R, Hochman Z, Dang YP (2018) Quantifying the economic impact of soil constraints on Australian agriculture: a case-study of wheat. Land Degrad Dev 29:3866–3875
Oyiga BC, Sharma RC, Baum M, Ogbonnaya FC, Léon J, Ballvora A (2018) Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant Cell Environ 41:919–935
Oyiga BC, Ogbonnaya FC, Sharma RC, Baum M, Léon J, Ballvora A (2019) Genetic and transcriptional variations in NRAMP-2 and OPAQUE1 genes are associated with salt stress response in wheat. Theor Appl Genet 132:323–346
Pang Y, Chen K, Wang X, Wang W, Xu J, Ali J, Li Z (2017) Simultaneous improvement and genetic dissection of salt tolerance of rice (Oryza sativa L.) by designed QTL pyramiding. Front Plant Sci 8:1275
Qin Y, Tian Y, Liu X (2015) A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. B Biochem Biophys Res Commun 464:428–433
Quamruzzaman M, Manik SMN, Shabala S, Zhou M (2021) Improving performance of salt-grown crops by exogenous application of plant growth regulators. Biomolecules 11:788
Rengasamy P (2006) World salinization with emphasis on Australia. J Exp Bot 57:1017–1023
Sbei H, Sato K, Shehzad T, Harrabi M, Okuno K (2014) Detection of QTLs for salt tolerance in Asian barley (Hordeum vulgare L.) by association analysis with SNP markers. Breed Sci 64:378–388
Shabala S, Munns R (2012) Salinity stress: physiological constraints and adaptive mechanisms. Plant Stress Physiol 1:59–93
Shabala S, Bose J, Hedrich R (2014) Salt bladders: do they matter? Trends Plant Sci 19:687–691
Shen Y, Shen L, Shen Z, Jing W, Ge H, Zhao J, Zhang W (2015) The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ 38:2766–2779
Sui N, Tian S, Wang W, Wang M, Fan H (2017) Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front Plant Sci 8:1337
USDA (2021) World Agricultural Production. Foreign Agricultural Service. WAP 03–21, March 2021. United States Department of Agriculture
Véry A-A, Nieves-Cordones M, Daly M, Khan I, Fizames C, Sentenac H (2014) Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J Plant Physiol 171:748–769
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L (2014) Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang X, Xu Y, Hu Z, Xu C (2018) Genomic selection methods for crop improvement: current status and prospects. Crop J 6:330–340
Wu H, Shabala L, Barry K, Zhou M, Shabala S (2013) Ability of leaf mesophyll to retain potassium correlates with salinity tolerance in wheat and barley. Physiol Plant 149:515–527
Xu Y-F, An D-G, Liu D-C, Zhang A-M, Xu H-X, Li B (2012b) Mapping QTLs with epistatic effects and QTL× treatment interactions for salt tolerance at seedling stage of wheat. Euphytica 186:233–245
Xu R, Wang J, Li C, Johnson P, Lu C, Zhou M (2012) A single locus is responsible for salinity tolerance in a Chinese landrace barley (Hordeum vulgare L). PLoS One 7:e43079
Xu B, Hrmova M, Gilliham M (2020) High affinity Na+ transport by wheat HKT1;5 is blocked by K+. Plant Direct 4:e00275
Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G, Cai J, Wu T, Moran N, Yu L (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166:945–959
Yarra R, Kirti P (2019) Expressing class I wheat NHX (TaNHX2) gene in eggplant (Solanum melongena L.) improves plant performance under saline condition. Funct Integr Genomics 19:541–554
Yarra R, He S-J, Abbagani S, Ma B, Bulle M, Zhang W-K (2012) Overexpression of a wheat Na+/H+ antiporter gene (TaNHX2) enhances tolerance to salt stress in transgenic tomato plants (Solanum lycopersicum L.). Plant Cell Tissue Organ Cult 111:49–57
Yarra R (2019) The wheat NHX gene family: potential role in improving salinity stress tolerance of plants. Plant Gene 18:100178
Yin L, Zhang H, Tang Z, Xu J, Yin D, Zhang Z, Yuan X, Zhu M, Zhao S, Li X (2021) rmvp: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom Proteom Bioinform. https://doi.org/10.1016/j.gpb.2020.10.007
Yousfi F-E, Makhloufi E, Marande W, Ghorbel AW, Bouzayen M, Berges H (2017) Comparative analysis of WRKY genes potentially involved in salt stress responses in Triticum turgidum L. ssp. durum. Front Plant Sci 7:2034
Zhao Y, Ai X, Wang M, Xiao L, Xia G (2016) A putative pyruvate transporter TaBASS2 positively regulates salinity tolerance in wheat via modulation of ABI4 expression. BMC Plant Biol 16:1–12
Zhao Z, Zhang G, Zhou S, Ren Y, Wang W (2017) The improvement of salt tolerance in transgenic tobacco by overexpression of wheat F-box gene TaFBA1. Plant Sci 259:71–85
Zhao C, Zhang H, Song C, Zhu J-K, Shabala S (2020) Mechanisms of plant responses and adaptation to soil salinity. Innovation 1:100017
Zhu M, Shabala S, Shabala L, Fan Y, Zhou M (2015) Evaluating predictive values of various physiological indices for salinity stress tolerance in wheat. J Agron Crop Sci 202:115–124
Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, Munns R, Shabala S (2016) Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J Exp Bot 67:835–844
Acknowledgements
The authors would like to thank Mr Mark Livermore at the Tasmanian Institute of Agriculture, University of Tasmania for English editing.
Funding
This research was funded by the National Natural Science Foundation of China (31871535) and the Grains Research and Development Corporation (GRDC) of Australia.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Human and animal rights
This study does not include human or animal subjects.
Additional information
Communicated by Aimin Zhang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Quamruzzaman, M., Manik, S.M.N., Shabala, S. et al. Genome-wide association study reveals a genomic region on 5AL for salinity tolerance in wheat. Theor Appl Genet 135, 709–721 (2022). https://doi.org/10.1007/s00122-021-03996-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03996-8