Abstract
Key message
A major quantitative trait locus (QTL) modulating soybean (Glycine max) branch angle was identified by linkage analysis using two bi-parental mapping populations with and without pedigree from wild soybean (Glycine soja).
Abstract
Soybean branch angle is a critical architectural trait that affects many other traits of agronomic importance associated with the plant’s productivity and grain yield and is thus a vital consideration in soybean breeding. However, the genetic basis for modulating this important trait in soybean and many other crops remain unknown. Previously, we developed a recombinant inbred line (RIL) population derived from a cross between a domesticated soybean (Glycine max) variety, Williams 82, and a wild soybean (Glycine soja) accession, PI 479,752, and observed drastic variation in plant architecture including branch angle among individual RILs. In this study, one of the RILs possessing extremely wide branch angle (WBA) was crossed with an elite soybean cultivar (LD00-3309) possessing narrow branch angle (NBA) to produce an F2 population composed of 147 plants and F2-derived F3 families for inheritance analysis and QTL mapping. We found that branch angle is controlled by a major QTL located on chromosome 19, designated qGmBa1 and that WBA—derived from the wild soybean accession—is dominant over NBA. This locus was also detected as a major one underlying branch angle by QTL mapping using a subset of the soybean nested association mapping (SoyNAM) population composed of 140 RILs, which were derived from a cross between a landrace, PI 437169B, possessing WBA and an elite variety, IA3023, possessing NBA. Molecular markers located in the QTL region defined by both mapping populations can be used for marker-assisted selection of branch angle in soybean breeding.



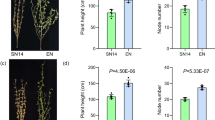
taken from 20 randomly selected individuals from the individual F3 families and merged together with 95% transparency

Similar content being viewed by others
References
Agudamu TY, Tatsuhiko S (2016) Branch development responses to planting density and yield stability in soybean cultivars. Plant Prod Sci 19(3):331–339
Asanome N, Ikeda T (1998) Effect of branch directions ° arrangement on soybean yield and yield components. J Agron Crop Sci 181:95–102
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Carter TE, Jr, Nelson RL, Sneller CH, Cui Z (2004) Genetic diversity in soybean. In: Shibles RM, Harper JE, Wilson RF, Shoemaker R (Eds)., Soybeans: improvement, production, and uses
Cocuron JC, Lrerouxel O, Drakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA 104:8550–8555
Dzievit MJ, Li X, Yu J (2018) Dissection of leaf angle variation in maize through genetic mapping and meta-analysis. Plant Genome 12:1800244
Food and Agricultural Organization of the United Nations (FAO) (2018) Value of agricultural production. Available at http://www.fao.org/faostat/en/#data/QV.
Foroutan-pour K, Dutilleul P, Smith DL (1999) Soybean canopy development as affected by population density and intercropping with corn: fractal analysis in comparison with other quantitative approaches. Crop Sci 39:1784–1791
Gille S, Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M (2011) O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23:4041–4053
Gunl M, Neumetzler L, Kraemer F, Souza A, Schultnik A, Pena M, Ws Y, Pauly M (2011) AXY8 encodes an α-fucoside, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23:4025–4040
Harder D, Sprague C, Renner K (2007) Effect of soybean row width and population on weeds, crop yield, and economic return. Weed Technol 21:744–752
Hsieh WY, Liao JC, Chang CY, Harrison T, Boucher C, Hsieh MH (2015) The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in Arabidopsis. Plant Physiol 168:490–501
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103:16666–16671
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Li J (2010) Regulation of osmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Khush G (2013) Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breed 132:143–146
Kim MY, Van K, Kang YJ, Kim KH, Lee S-H (2012) Tracing soybean domestication history: from nucleotide to genome. Breed Sci 61:445–452
Kong Y, Zhou G, Yin Y, Xu Y, Pattathil S, Hahn MG (2011) Molecular analysis of a family of Arabidopsis genes related to galacturonosyltransferases. Plant Physiol 155:1791–1805
Ku LX, Wei XM, Zhang SF, Zhang J, Guo SL, Chen Y (2011) Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L). PLOS One 6:e20621
Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J (2007) LAZY1 Controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17:402–410
Lu M, Zhou F, Xie CX, Li MS, Xu YB, Marilyn W, Zhang SH (2007) Construction of a SSR linkage map and mapping of quantitative trait loci (QTL) for leaf angle and leaf orientation with an elite maize hybrid. Hereditas 29:1131–1138
Mace ES, Buhariwalla KK, Buhariwalla HK, Crouch JH (2003) A High-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Mol Biol Rep 21:459–460
Mantilla-Perez MB, Fernandez MGS (2017) Differential manipulation of leaf angle throughout the canopy: current status and prospects. J Exp Bot 68:5699–5717
Norman JM, Cambell GS (1989) Canopy Structure. In: Pearcy RW, Ehleringer JR, Mooney HA, Rundel PW (eds) Plant physiol ecol. Springer, Dordrecht
Pendleton JWS, GE, Winter SR, Johnston TJ, (1968) Field investigations of the relationships of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis. Agron J 60:422–424
Peng M, Cui Y, Bi YM, Rothstein SJ (2006) AtMBD9: A Protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. Plant J 46:282–296
Reinhardt D, Kuhlemeier C (2002) Plant architecture. EMBO Rep 3:846–851
Schon MK, Blevins DG (1990) Foliar boron applications increase the final number of branches and pods on branches of field-grown soybeans. Plant Physiol 92:602–607
Settimi JR, Board JE (1988) Photoperiod and planting date effects on the spatial distribution of branch development in soybean. Crop Sci 28:259–263
Swarm SA, Sun L, Wang X, Wang W, Brown PJ, Ma J, Nelson RL (2019) Genetic dissection of domestication-related traits in soybean through genotyping-by-sequencing of two interspecific mapping populations. Theor Appl Genet 132:1195–1209
Tian J, Wang C, Xia J, Wu L, Xu G, Wu W, Li D, Qin W, Han X, Chen Q, Jin W, Tian F (2019) Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365:658–654
Tollenaar M, Lee EA (2002) Yield potential, yield stability and stress tolerance in maize. Field Crops Res 75:161–169
Truong SK, McCormick RF, Rooney WL, Mullet JE (2015) Harnessing genetic variation in leaf angle to increase productivity of Sorghum Bicolor. Genetics 201:1229–1238
Virdi KS, Sreekanta S, Dobbels A, Haaning A, Jarquin D, Stupar RM, Lorenz AJ, Muehlbauer GJ (2021) Branch angle and leaflet shape are associated with canopy coverage in soybean. Sci Rep. https://doi.org/10.21203/rs.3.rs-806530/v1
Wang X, Chen L, Ma J (2019) Genomic introgression through interspecific hybridization counteracts genetic bottleneck during soybean domestication. Genome Biol 20:22
Weaver DB, Akridge RL, Thomas CA (1991) Growth habit, planting date, and row spacing effects on late planted soybean. Crop Sci 31:805–810
Wu X, Tang D, Li M, Wang K, Cheng Z (2013) Loose plant architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161:317–329
Xavier A, Hall B, Hearst AA, Cherkauer KA, Rainey KM (2017) Genetic architecture of phenomic-enabled canopy coverage in Glycine max. Genetics 206:1081–1089
Yang L, Wu G, Poethig RS (2012) Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA mediated translational repression in Arabidopsis. Proc Natl Acad Sci USA 109:315–320
Yu B, Lin Z, Li H, Li X, Li J, Wang Y, Zhang X, Zhu Z, Zhai W, Wang X, Xie D, Sun C (2007) TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J 52:891–898
Zhang J, Ku LX, Han ZP, Guo SL, Liu HJ, Zhang ZZ, Cao LR, Cui XJ, Chen YH (2014) The ZmCLA4 gene in the qLA4-1 QTL controls leaf angle in maize (Zea mays L.). J Exp Bot 65:5063–5076
Zhang N, Yu H, Yu H, Cai Y, Huang L, Xu C, Xiong G, Meng X, Wang J, Chen H, Liu G, Jing Y, Yuan Y, Liang Y, Li S, Smith SM, Li J, Wang Y (2018) A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of Auxin. Plant Cell 30:1461–1475
Zhu XG, Long SP, Ort DR (2008) What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr Opin Biotech 19:153–159
Acknowledgments
We would like to thank Xutong Wang, Jingbo Duan, and Liyang Chen for help with harvesting soybean plants.
Funding
This work was mainly supported by (# 2015–67013-22811, 2018–67013-27425) funded by the Agriculture and Food Research Initiative of the USDA National Institute of Food and Agriculture, and partially supported by Indiana Soybean Alliance.
Author information
Authors and Affiliations
Contributions
JM designed research, CBC, WW, YW, GJF, ZW, and BR performed research, CBC, DW, and BR analyzed data, CBC and JM wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Data availability
All data presented in this manuscript are included in the supplemental tables. All materials are available to the public upon request and under material transfer agreement.
Additional information
Communicated by Istvan Rajcan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Clark, C.B., Wang, W., Wang, Y. et al. Identification and molecular mapping of a major quantitative trait locus underlying branch angle in soybean. Theor Appl Genet 135, 777–784 (2022). https://doi.org/10.1007/s00122-021-03995-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03995-9