Abstract
Key message
We constructed a homoeologous recombination-based bin map of wheat chromosome 7B, providing a unique physical framework for further study of chromosome 7B and its homoeologues in wheat and its relatives.
Abstract
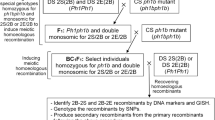
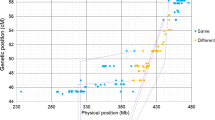
Homoeologous recombination leads to the dissection and diversification of the wheat genome. Advances in genome sequencing and genotyping have dramatically improved the efficacy and throughput of homoeologous recombination-based genome studies and alien introgression in wheat and its relatives. In this study, we aimed to physically dissect and map wheat chromosome 7B by inducing meiotic recombination of chromosome 7B with its homoeologues 7E in Thinopyrum elongatum and 7S in Aegilops speltoides. The special genotypes, which were double monosomic for chromosomes 7B’ + 7E’ or 7B’ + 7S’ and homozygous for the ph1b mutant, were produced to enhance 7B − 7E and 7B − 7S recombination. Chromosome-specific DNA markers were developed and used to pre-screen the large recombination populations for 7B − 7E and 7B − 7S recombinants. The DNA marker-mediated preselections were verified by fluorescent genomic in situ hybridization (GISH). In total, 29 7B − 7E and 61 7B − 7S recombinants and multiple chromosome aberrations were recovered and delineated by GISH and the wheat 90 K SNP assay. Integrated GISH and SNP analysis of the recombinants physically mapped the recombination breakpoints and partitioned wheat chromosome 7B into 44 bins with 523 SNPs assigned within. A composite bin map was constructed for chromosome 7B, showing the bin size and physical distribution of SNPs. This provides a unique physical framework for further study of chromosome 7B and its homoeologues. In addition, the 7B − 7E and 7B − 7S recombinants extend the genetic variability of wheat chromosome 7B and represent useful germplasm for wheat breeding. Thereby, this genomics-enabled chromosome engineering approach facilitates wheat genome study and enriches the gene pool of wheat improvement.







Similar content being viewed by others
References
Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale L, Mascher M, Spannagl M, Wiebe K, Jordan KW, Golan G, Deek J, Ben-Zvi G, Himmelbach A, MacLachlan RP, Sharpe AG, Fritz A, Ben-David R, Budak H, Fahima T, Korol A, Faris JD, Hernandez A, Mikel MA, Levy AA, Steffenson B, Maccaferri M, Tuberosa R, Cattivelli L, Faccioli P, Ceriotti A, Kashkush K, Pourkheirandish M, Komatsuda T, Eilam T, Sela H, Sharon A, Ohad N, Chamovitz DA, Mayer KFX, Stein N, Ronon G, Peleg Z, Pozniak CJ, Akhunov ED, Distelfeld A (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357:93–97
Cai X, Jones S, Murray T (1998) Molecular cytogenetic characterization of Thinopyrum and wheat–Thinopyrum translocated chromosomes in a wheat Thinopyrum amphiploid. Chromosome Res 6:185–189
Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ (1995) Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128
Chen P, Liu W, Yuan J, Wang X, Zhou B, Wang S, Zhang S, Feng Y, Yang B, Liu G, Liu D, Qi L, Zhang P, Friebe B, Gill BS (2005) Development and characterization of wheat-Leymus racemosus translocation lines with resistance to Fusarium Head Blight. Theor Appl Genet 111:941–948
Crasta OR, Francki MG, Bucholtz DB, Sharma HC, Zhang J, Wang RC, Ohm HW, Anderson JM (2000) Identification and characterization of wheat-wheatgrass translocation lines and localization of barley yellow dwarf virus resistance. Genome 43:698–706
Dai K, Zhao R, Shi M, Xiao J, Yu Z, Jia Q, Wang Z, Yuan C, Sun H, Cao A, Zhang R, Chen P, Li Y, Wang H, Wang X (2020) Dissection and cytological mapping of chromosome arm 4VS by the development of wheat-Haynaldia villosa structural aberration library. Theor Appl Genet 133:217–226
Danilova TV, Friebe B, Gill BS (2014) Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet 127:715–730
Danilova TV, Zhang G, Liu W, Friebe B, Gill BS (2017) Homoeologous recombination-based transfer and molecular cytogenetic mapping of a wheat streak mosaic virus and Triticum mosaic virus resistance gene Wsm3 from Thinopyrum intermedium to wheat. Theor Appl Genet 130:549–556
Danilova TV, Friebe B, Gill BS (2019) Production of a complete set of wheat-barley group-7 chromosome recombinants with increased grain β-glucan content. Theor Appl Genet 132:3129–3141
Devos KM, Dubcovsky J, Dvorak J, Chinoy CN, Gale MD (1995) Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor Appl Genet 91:282–288
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Faris JD (2014) Wheat domestication: key to agricultural revolutions past and future. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources. Vol. 1. Managing, sequencing and mining genetic resources. Springer, Netherlands, pp 439–464
Feldman M, Levy AA (2012) Genome evolution due to allopolyploidization in wheat. Genetics 192:763–774
Friebe B, Jiang J, Raupp WJ, Mclntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87
Friebe B, Zhang P, Linc G, Gill BS (2005) Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet Genome Res 109:293–297
Friebe B, Qi L, Liu C, Gill B (2011) Genetic compensation abilities of Aegilops speltoides chromosomes for homoeologous B-genome chromosomes of polyploid wheat in disomic S(B) chromosome substitution lines. Cytogenet Genome Res 134:144–150
Gill KS, Gill BS (1991) A DNA fragment mapped within the submicroscopic deletion of Ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics 129:257–259
Gyawali Y, Zhang W, Chao S, Xu SS, Cai X (2019) Delimitation of wheat ph1b deletion and development of the ph1b-specific DNA markers. Theor Appl Genet 132:195–204
Grewal S, Hubbart-Edwards S, Yang C, Devi U, Baker L, Heath J, Scholefield SAD, Howells C, Yarde J, Isaac P, King IP, King J (2020) Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant Biotechnol J 18:743–755
International Wheat Genome Sequencing Consortium (IWGSC) (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:6403
King J, Grewal S, Yang CY, Hubbart Edwards S, Scholefield D, Ashling S, Harper JA, Allen AM, Edwards KJ, Burridge AJ, King IP (2018) Introgression of Aegilops speltoides segments in Triticum aestivum and the effect of the gametocidal genes. Ann Bot 121:229–240
Li H, Dong Z, Ma C, Tian X, Qi Z, Wu N, Friebe B, Xiang Z, Xia Q, Liu W, Li T (2019) Physical mapping of stem rust resistance gene Sr52 from Dasypyrum villosum based on ph1b-induced homoeologous recombination. Int J Mol Sci 20:4887
Ling H, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, Yu Y, Du H, Qi M, Li Y, Lu H, Yu H, Cui Y, Wang N, Chen C, Wu H, Zhao Y, Zhang J, Li Y, Zhou W, Zhang B, Hu W, van Eijk MJT, Tang J, Witsenboer HMA, Zhao S, Li Z, Zhang A, Wang D, Liang C (2018) Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557:424–428
Liu W, Danilova TV, Rouse MN, Bowden RL, Friebe B, Gill BG (2013) Development and characterization of a compensating wheat-Thinopyrum intermedium Robertsonian translocation with Sr44 resistance to stem rust (Ug99). Theor Appl Genet 126:1167–1177
Long Y, Chao WS, Ma G, Xu SS, Qi L (2017) An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theor Appl Genet 130:597–607
Lukaszewski A, Rybka K, Korzun V, Malyshev S, Lapinski B, Whitkus R (2004) Genetic and physical mapping of homoeologous recombination points involving wheat chromosome 2B and rye chromosome 2R. Genome 47:36–45
Luo M, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo X, Zhu T, Wang L, Wang Y, McGuire PE, Liu S, Long H, Ramasamy RK, Rodriguez JC, Van SL, Yuan L, Wang Z, Xia Z, Xiao L, Anderson OD, Ouyang S, Liang Y, Zimin AV, Pertea G, Qi P, Bennetzen JL, Dai X, Dawson MW, Müller H, Kugler K, Rivarola-Duarte L, Spannagl M, Mayer KFX, Lu F, Becan MW, Lerou P, Li P, You FM, Sun Q, Liu Z, Lyons E, Wicher T, Zalzberg SL, Devos KM, Dvořák J (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551:498–502
Mago R, Spielmeyer W, Lawrence G, Lagudah E, Ellis J, Pryor A (2002) Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor Appl Genet 104:1317–1324
Miftahudin RK, Ross K, Ma XF, Mahmoud AA, Layton J, Milla MA, Chikmawati T, Ramalingam J, Feril O, Pathan MS, Momirovic GS, Kim S, Chema K, Fang P, Haule L, Struxness H, Birkes J, Yaghoubian C, Skinner R, McAllister J, Nguyen V, Qi LL, Echalier B, Gill BS, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvorák J, Dilbirligi M, Gill KS, Peng JH, Lapitan NL, Bermudez-Kandianis CE, Sorrells ME, Hossain KG, Kalavacharla V, Kianian SF, Lazo GR, Chao S, Anderson OD, Gonzalez-Hernandez J, Conley EJ, Anderson JA, Choi DW, Fenton RD, Close TJ, McGuire PE, Qualset CO, Nguyen HT, Gustafson JP (2004) Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168:651–663
Naranjo T, Fernández-Rueda P (1996) Pairing and recombination between individual chromosomes of wheat and rye in hybrids carrying the ph1b mutation. Theor Appl Genet 93:242–248
Neelam K, Brown-Guedira G, Huang L (2013) Development and validation of a breeder-friendly KASPar marker for wheat leaf rust resistance locus Lr21. Mol Breed 31:233–237
Niu Z, Klindworth DL, Friesen TL, Chao S, Jin Y, Cai X, Xu SS (2011) Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187:1011–1021
Niu Z, Klindworth DL, Yu G, Friesen TL, Chao S, Jin Y, Cai X, Ohm J-B, Rasmussen JB, Xu SS (2014) Development and characterization of wheat lines carrying stem rust resistance gene Sr43 derived from Thinopyrum ponticum. Theor Appl Genet 129:969–980
Patokar C, Sepsi A, Schwarzacher T, Kishii M, Heslop-Harrison JS (2016) Molecular cytogenetic characterization of novel wheat-Thinopyrum bessarabicum recombinant lines carrying intercalary translocations. Chromosoma 125:163–172
Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos-Korts D, Goué N, Balfourier F, Molnar-Lang M, Lage J, Kilian B, Ozkan H, Waite D, Dyer S, Alaux M, Letellier T, Russell J, Keller B, Eeuwijk F, Spannagl M, Mayer K, Waugh R, Stein N, Cattivelli L, Haberer G, Charmet G, Salse J (2019) Tracing the ancestry of modern bread wheats. Nat Genet 51:905–911
Qi L, Echalier B, Friebe B, Gill B (2003) Molecular characterization of a set of wheat deletion stocks for use in chromosome bin mapping of ESTs. Funct Integr Genom 3:39–55
Qi LL, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombina-tion, chromosome engineering and crop improvement. Chromo-some Res 15:3–19
Rabinovich S (1998) Importance of wheat-rye translocations for breeding modern cultivar of Triticum aestivum L. Euphytica 100:323–340
Rey M, Calderón M, Prieto P (2015) The use of the ph1b mutant to induce recombination between the chromosomes of wheat and barley. Front Plant Sci. https://doi.org/10.3389/fpls.2015.00160
Riley R, Chapman V (1958) Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182:713–715
Riley R, Chapman V, Kimber G (1959) Genetic control of chromosome pairing in intergeneric hybrids with wheat. Nature 185:1244–1246
Riley R, Chapman V, Johnson R (1968) Introduction of yellow rust resistance of Aegilops comosa into wheat by genetically induced homoeologous recombination. Nature 217:383–384
Roberts MA, Reader SM, Dalgliesh C, Miller TE, Foote TN, Fish LJ, Snape JW, Moore G (1999) Induction and characterization of Ph1 wheat mutants. Genetics 153:1909–1918
Šafář J, Šimková H, Kubaláková M, Číhalíková J, Suchánková P, Bartoš J, Doležel J (2010) Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet Genome Res 129:211–223
Schwarzacher T, Anamthawat-Jónsson K, Harrison GE, Islam AKMR, Jia JZ, King IP, Leitch AR, Miller TE, Reader SM, Rogers WJ, Shi M, Heslop-Harrison JS (1992) Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor Appl Genet 84:365–375
Sears ER (1954) The aneuploids of common wheat. Mo Agric Exp Stn Res Bull 572:1–59
Sears ER (1966) Nullisomic-tetrasomic combinations in hexaploid wheat. In: Riley R, Lewis KR (eds) Chromosome manipulation and plant genetics. Oliver and Boyd, Edinburgh, pp 29–45
Sears ER (1977) An induced mutant with homoeologous pairing in common wheat. Can J Genet Cytol 19:778–786
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic–physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genom 4:12–25
Tsujimoto H, Tsunewaki K (1988) Gametocidal genes in wheat and its relatives. III. Chromosome location and effects of two Aegilops speltoides-derived gametocidal genes in common wheat. Genome 30:239–244
Venske E, Santos RS, Busanello C, Gustafson P, Oliveira AC (2019) Bread wheat: a role model for plant domestication and breeding. Hereditas 156:16
Wang S, Wong D, Forrest K, Allen A, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, International Wheat Genome Sequencing Consortium, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang H, Sun S, Ge W, Zhao L, Hou B, Wang K, Lyu Z, Chen L, Xu S, Guo J, Li M, Su P, Li X, Wang G, Bo C, Fang X, Zhuang W, Cheng X, Wu J, Dong L, Chen W, Li W, Xiao G, Zhao J, Hao Y, Xu Y, Gao Y, Liu W, Liu Y, Yin H, Li J, Li X, Zhao Y, Wang X, Ni F, Ma X, Li A, Xu SS, Bai G, Nevo E, Gao C, Ohm H, Kong L (2020) Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368:eaba5435
Zhang R, Hou F, Feng Y, Zhang W, Zhang M, Chen P (2015) Characterization of a Triticum aestivum–Dasypyrum villosum T2VS 2DL translocation line expressing a longer spike and more kernels traits. Theor Appl Genet 128:2415–2425
Zhang W, Cao Y, Zhang M, Zhu X, Ren S, Long Y, Gyawali Y, Chao S, Xu S, Cai X (2017) Meiotic homoeologous recombination-based alien gene introgression in the genomics era of wheat. Crop Sci 57:1189–1198
Zhang W, Zhang M, Zhu X, Cao Y, Sun Q, Ma G, Chao S, Yan C, Xu S, Cai X (2018a) Molecular cytogenetic and genomic analyses reveal new insights into the origin of the wheat B genome. Theor Appl Genet 131:365–375
Zhang W, Zhu X, Zhang M, Chao S, Xu S, Cai X (2018b) Meiotic homoeologous recombination-based mapping of wheat chromosome 2B and its homoeologues in Aegilops speltoides and Thinopyrum elongatum. Theor Appl Genet 131:2381–2395
Zhang W, Zhu X, Zhang M, Shi G, Liu Z, Cai X (2019) Chromosome engineering-mediated introgression and molecular mapping of novel Aegilops speltoides-derived resistance genes for tan spot and Septoria nodorum blotch diseases in wheat. Theor Appl Genet 132:2605–2614
Zhang M, Zhang W, Zhu X, Sun Q, Chao S, Yan C, Xu S, Fiedler J, Cai X (2020) Partitioning and physical mapping of wheat chromosome 3B and its homoeologue 3E in Thinopyrum elongatum by inducing homoeologous recombination. Theor Appl Genet 133:1277–1289
Zhao R, Wang H, Xiao J, Bie T, Cheng S, Jia Q, Yuan C, Zhang R, Cao A, Chen P, Wang X (2013) Induction of 4VS chromosome recombinants using the CS ph1b mutant and mapping of the wheat yellow mosaic virus resistance gene from Haynaldia villosa. Theor Appl Genet 126:2921–2930
Acknowledgements
We thank Mary Osenga for her help in high-throughput SNP genotyping and Drs. Lili Qi and Rebekah Oliver for their critical review of the manuscript. This project has been supported by Agriculture and Food Research Initiative Competitive Grant No. 2013-67013-21121 and 2019-67013-29164 from the USDA National Institute of Food and Agriculture.
Author information
Authors and Affiliations
Contributions
M.Z. contributed to recombinant production and analysis, SNP assays, GISH, molecular marker development and analysis, and chromosome mapping, and was involved in data analysis and manuscript preparation. W.Z. and X.Z. participated in crossing, chromosome-specific marker analysis, and GISH. Q.S. and C.Y. analyzed DNA sequence data. J.F. ran high-throughput SNP assays. S.S.X. was involved in manuscript preparation. X.C. designed and coordinated this work, and was involved in crosses, data analysis and interpretation, and manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by P. Heslop-Harrison.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, M., Zhang, W., Zhu, X. et al. Dissection and physical mapping of wheat chromosome 7B by inducing meiotic recombination with its homoeologues in Aegilops speltoides and Thinopyrum elongatum. Theor Appl Genet 133, 3455–3467 (2020). https://doi.org/10.1007/s00122-020-03680-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03680-3