Abstract
Key message
A CIho 5791 × Tifang recombinant inbred mapping population was developed and used to identify major dominant resistance genes on barley chromosomes 6H and 3H in CI5791 and on 3H in Tifang.
Abstract
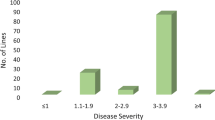
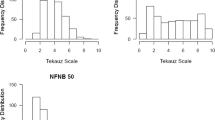
The barley line CIho 5791 confers high levels of resistance to Pyrenophora teres f. teres, causal agent of net form net blotch (NFNB), with few documented isolates overcoming this resistance. Tifang barley also harbors resistance to P. teres f. teres which was previously shown to localize to barley chromosome 3H. A CIho 5791 × Tifang F6 recombinant inbred line (RIL) population was developed using single seed descent. The Illumina iSelect SNP platform was used to identify 2562 single nucleotide polymorphism (SNP) markers across the barley genome, resulting in seven linkage maps, one for each barley chromosome. The CIho 5791 × Tifang RIL population was evaluated for NFNB resistance using nine P. teres f. teres isolates collected globally. Tifang was resistant to four of the isolates tested whereas CIho 5791 was highly resistant to all nine isolates. QTL analysis indicated that the CIho 5791 resistance mapped to chromosome 6H whereas the Tifang resistance mapped to chromosome 3H. Additionally, CIho 5791 also harbored resistance to two Japanese isolates that mapped to a 3H region similar to that of Tifang. SNP markers and RILs harboring both 3H and 6H resistance will be useful in resistance breeding against NFNB.



Similar content being viewed by others
References
Abu Qamar M, Liu ZH, Faris JD, Chao S, Edwards MC, Lai Z, Franckowiak JD, Friesen TL (2008) A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor Appl Genet 117:1261–1270
Arabi MI, Sarrafi A, Barrault G, Albertini L (1990) Inheritance of partial resistance to net blotch in barley. Plant Breed 105:150–155
Bockelman HE, Sharp EL, Eslick RF (1977) Trisomic analysis of genes for resistance to scald and net blotch in several barley cultivars. Can J Bot 55:2142–2148
Cantalapiedra CP, Boudiar R, Casas AM, Igartua E, Contreras-Moreira B (2015) Barleymap: physical and genetic mapping of nucleotide sequences and annotation loci in barley. Mol Breeding 35:1–11
Ciuffetti LM, Manning VA, Pandelova I, Betts MF, Martinez JP (2010) Host-selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis-wheat interaction. New Phytol 187:911–919
Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, Hedley P, Tondelli A, Pecchioni N, Francia E, Korzun V, Walther A, Waugh R (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44:1388–1392
Cromey MG, Parkes RA (2003) Pathogenic variation in Drechslera teres in New Zealand. N Z Plant Protect 56:251–256
Douglas GB, Gordon LL (1985) Quantitative genetics of net blotch resistance in barley. N Z J Agric Res 28:157–164
Friesen TL, Faris JD, Lai Z, Steffenson BJ (2006) Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled-haploid barley population. Genome 49:855–859
Geschele EE (1928) The response of barley to parasitic fungi Helminthosporium teres Sacc. Bull Appl Bot Genet Plant Breed 19:371–384 (in Rev Appl Mycol 8:165)
Graner A, Foroughi-Wehr B, Tekauz A (1996) RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica 91:229–234
Gray GG (1966) Genetic systems in the net blotch disease complex of barley. Phd Dissertation. North Dakota State University, Fargo, ND
Gupta S, Loughman R (2001) Current virulence of Pyrenophora teres on barley in Western Australia. Plant Dis 85:960–966
Gupta S, Wielinga C, Li C, Cakir M, Platz G, Loughman R, Lance R, Appels R (2004) Gene distribution and SSR markers linked with net type net blotch resistance in barley. In: Proceedings of the 9th International Barely Genetics Symposium (Spunar J, Janikova J, eds) Agricultural Research Institute Kromeriz Ltd., Brno, Czech Republic, June 20–26. p 668–673
Ho KM, Tekauz A, Choo TM, Martin RA (1996) Genetic studies on net blotch resistance in a barley cross. Can J Plant Sci 76:715–720
Illumina Inc (2010) Whole genome genotyping with the Infinium assay. Illumina, San Diego
Jalli M (2004) Suitability of a selected barley differential set for Pyrenophora teres f. teres virulence screening. In: Proceedings of the 9th International Barely Genetics Symposium (Spunar J, Janikova J, eds) Agricultural Research Institute Kromeriz, Ltd., Brno, Czech Republic, June 20–26 p 266–269
Jalli M, Robinson J (2000) Stable resistance in barley to Pyrenophora teres f. teres isolates from the Nordic-Baltic region after increase on standard host genotypes. Euphytica 113:71–77
Joehanes R, Nelson JC (2008) QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 24:2788–2789
Jonsson R, Bryngelsson T, Gustafsson M (1997) Virulence studies of Swedish net blotch isolates (Drechslera teres) and identification of resistant barley lines. Euphytica 94:209–218
Khan TN, Boyd WJR (1969a) Physiologic specialization in Drechslera teres. Aust J Biol Sci 22:1229–1235
Khan TN, Boyd WJR (1969b) Inheritance of resistance to net blotch in barley. II. Genes conditioning resistance against race WA-2. Can J Genet Cytol 11:592–597
Khan TN, Boyd WJR (1971) Genetics of host resistance to net blotch in Barley. In: Nilan RA (ed) Barley Genetics II. Washington State University Press, Pullman, pp 484–492
Liu Z, Ellwood SR, Oliver RP, Friesen TL (2011) Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol Plant Pathol 12:1–19
Liu Z, Zhang Z, Faris JD, Oliver RP, Syme R, McDonald MC, McDonald BM, Solomon PS, Lu S, Shelver WL, Xu S, Friesen TL (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog 8:e1002467
Liu Z, Holmes DJ, Faris JD, Chao S, Brueggeman RS, Edwards MC, Friesen TL (2015) Necrotrophic effector‐triggered susceptibility (NETS) underlies the barley–Pyrenophora teres f. teres interaction specific to chromosome 6H. Mol Plant Pathol 16:188–200
Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng S-C, Maier CS, Wolpert TJ (2012) Tricking the guard: exploiting plant defense for disease susceptibility. Science 338:659–662
Lorieux M (2012) MapDisto: fast and efficient computation of genetic linkage maps. Mol Breed 30:1231–1235
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Munoz-Amatriain M, Close TJ, Wise RP, Schulman AH, Himmelbach A, Mayer KFX, Scholz U, Poland JA, Stein N, Waugh R (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76(718):727
Mathre DE (1997) Compendium of barley diseases, 2nd edn. American Phytopathological Society. APS Press, St. Paul
McDonald WC, Buchannon KW (1962) The inheritance of variability in Pyrenophora teres. Barley Newsl 6:40
Mode CJ, Schaller CW (1958) Two additional factors for host resistance to net blotch in barley. Agron J 50:15–18
Raman H, Platz GJ, Chalmers KJ, Raman R, Read BJ, Barr AR, Moody DB (2003) Mapping of genetic regions associated with net form of net blotch resistance in barley. Aust J Agric Res 54:1359–1367
Richards J, Chao S, Friesen T, Brueggeman R (2016) Fine mapping of the barley chromosome 6H net form net blotch susceptibility locus. G3 (Bethesda) 6(7):1809–1818. doi:10.1534/g3.116.028902
SAS Institute Inc (2013) 12.3 User’s guide: high-performance procedures. SAS Institute, Cary
Schaller CW (1955) Inheritance of resistance to net blotch of barley. Phytopathology 45:174–176
Shipton WA, Khan TN, Boyd WJR (1973) Net blotch of barley. Rev Plant Pathol 52:269–290
Shjerve RA, Faris JD, Brueggeman RS, Yan C, Zhu Y, Koladia V, Friesen TL (2014) Evaluation of a Pyrenophora teres f. teres mapping population reveals multiple independent interactions with the barley 6H chromosome region. Fungal Genet Biol 70:104–112
Steffenson BJ, Webster RK (1992) Quantitative resistance to Pyrenophora teres f. teres in barley. Phytopathology 82:407–411
Tekauz A (1985) A numerical scale to classify reactions of barley to Pyrenophora teres. Can J Plant Pathol 7:181–183
Tekauz A (1990) Characterization and distribution of pathogenic variation in Pyrenophora teres f. teres and P. teres f. maculata from western Canada. Can J Plant Pathol 12:141–148
Wu H-L, Steffenson BJ, Zhong S (2003) Genetic variation for virulence and RFLP markers in Pyrenophora teres. Can J Plant Pathol 25:82–90
Yun SJ, Gyenis L, Hayes PM, Matus I, Smith KP, Steffenson BJ, Muehlbauer GJ (2005) Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci 45:2563–2572
Acknowledgments
The authors would like to thank Danielle Holmes for technical assistance. This research was supported by funding from USDA-ARS CRIS Project 5442-22000-048-00D, USDA-NIFA-AFRI grant#2011-68002-30029 (T-CAP), the North Dakota Barley Council, and the Montana Wheat and Barley Committee. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflicts of interest for this article.
Ethical standards
All experiments performed complied with the ethical standards of the USDA-ARS and North Dakota State University.
Additional information
Communicated by F. Ordon.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koladia, V.M., Faris, J.D., Richards, J.K. et al. Genetic analysis of net form net blotch resistance in barley lines CIho 5791 and Tifang against a global collection of P. teres f. teres isolates. Theor Appl Genet 130, 163–173 (2017). https://doi.org/10.1007/s00122-016-2801-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2801-4