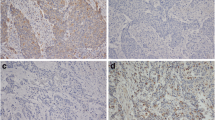
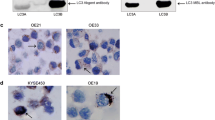
Abstract
Less than 15% of patients with esophageal squamous cell carcinoma (ESCC) survive 5 years after diagnosis. A better understanding of the biology of these tumors and the development of clinical biomarkers is needed. Autophagy is a physiological mechanism involved in the turnover of cellular components that plays a key role in cancer. This study evaluated the differential levels of three key regulators of autophagy (SQSTM1, MAP1LC3B, and BECN1) in patients with ESCC, associating autophagy with histopathologic features, including the grade of differentiation, mitotic rate, inflammation score, and the intensity of tumor-infiltrating lymphocytes. Nuclear morphometry of the tumor parenchyma was also assessed, associating it with autophagy and histopathology. All three markers significantly increased in patients with ESCC compared to the control group. Based on the mean expression of each protein in the control group, 57% of patients with ESCC had high levels of all three markers compared to control patients (14%). The most frequent profiles found in ESCC were BECNhigh/MAP1LC3high and BECNhigh/SQSTM1high. According to the TCGA database, we found that the main autophagy genes were upregulated in ESCC. Moreover, high levels of autophagy markers were associated with a poor prognosis. Considering nuclear morphometry, ESCC samples showed a significant reduction in nuclear area, which was strongly negatively correlated with autophagy. Finally, the percentage of normal nuclei was associated with tumor differentiation, while poorly differentiated tumors showed lower SQSTM1 levels. ESCC progression may involve increased autophagy and changes in nuclear structure, associated with clinically relevant histopathological features.
Key messages
-
Autophagy markers are co-increased in primary ESCC.
-
Autophagy negatively correlates with nuclear morphometry in ESCC parenchyma.
-
Autophagy and nuclear morphometry are associated with histopathological features.
-
Autophagy is increased in ESCC-TCGA database and associated with poor prognosis.




Similar content being viewed by others
Data availability
Immunohistochemistry generated in this study is not publicly available due to ethical privacy, but can be obtained from the corresponding author upon reasonable request.
Abbreviations
- AutoIndex:
-
Autophagic Index
- ATG:
-
Autophagy genes or proteins
- BECN1:
-
Beclin-1
- ESCC:
-
Esophageal squamous cell carcinoma
- EC:
-
Esophageal carcinoma
- MAP1LC3A/B/C (LC3A/B/C):
-
Microtubule-associated protein 1 light chain 3 (isoforms A, B, and C)
- NMA:
-
Nuclear morphometric analysis
- tNMA:
-
Nuclear morphometric analysis in tissue
- NII:
-
Nuclear irregularity index
- N nuclei:
-
Normal nuclei
- OST:
-
Overall survival time
- SQSTM1:
-
Sequestosome 1
- SR nuclei:
-
Small and regular nuclei
- TCGA:
-
The Cancer Genome Atlas
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72:7–33
Fagundes RB, de Carli D, Xaubet RV, Cantarelli JC (2016) Unchanging pattern of prevalence of esophageal cancer, overall and by histological subtype, in the endoscopy service of the main referral hospital in the central region of Rio Grande do Sul State, in Southern Brazil. Dis Esophagus 29:603–606
Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V (2020) Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol 13:1010–1021
Murphy G, McCormack V, Abedi-Ardekani B et al (2017) International cancer seminars: a focus on esophageal squamous cell carcinoma. Ann Oncol 28:2086–2093
Galluzzi L, Pietrocola F, Bravo-San Pedro JM et al (2015) Autophagy in malignant transformation and cancer progression. EMBO J 34:856–880
Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147:728–741
Yu L, Chen Y, Tooze SA (2018) Autophagy pathway: cellular and molecular mechanisms. Autophagy 14:207–215
Li X, He S, Ma B (2020) Autophagy and autophagy-related proteins in cancer. Mol Cancer 19:12
Cao Y, Klionsky DJ (2007) Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res 17:839–849
Kang R, Zeh HJ, Lotze MT, Tang D (2011) The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18:571–580
He H, Dang Y, Dai F et al (2003) Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 278:29278–29287
Kabeya Y, Mizushima N, Ueno T et al (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728
Pankiv S, Clausen TH, Lamark T et al (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145
Sahani MH, Itakura E, Mizushima N (2014) Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 10:431–441
Murrow L, Debnath J (2013) Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol 8:105–137
Ryter SW, Cloonan SM, Choi AM (2013) Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol Cells 36:7–16
Vessoni AT, Filippi-Chiela EC, Menck CFM, Lenz G (2013) Autophagy and genomic integrity. Cell Death Differ 20:1444–1454
Amaravadi R, Kimmelman AC, White E (2016) Recent insights into the function of autophagy in cancer. Genes Dev 30:1913–1930
Degenhardt K, Mathew R, Beaudoin B et al (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10:51–64
Poillet-Perez L, Sharp DW, Yang Y et al (2020) Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nat Cancer 1:923–934
Bustos SO, Antunes F, Rangel MC, Chammas R (2020) Emerging autophagy functions shape the tumor microenvironment and play a role in cancer progression - implications for cancer therapy. Front Oncol 10:606436
Janji B, Berchem G, Chouaib S (2018) Targeting autophagy in the tumor microenvironment: new challenges and opportunities for regulating tumor immunity. Front Immunol 9:887
Das CK, Mandal M, Kögel D (2018) Pro-survival autophagy and cancer cell resistance to therapy. Cancer Metastasis Rev 37:749–766
O’Donovan TR, O’Sullivan GC, McKenna SL (2011) Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy 7:509–524
Cao D, Shan D, Yan W et al (2021) Chaperone-mediated autophagy affects tumor cell proliferation and cisplatin resistance in esophageal squamous cell carcinoma. Thorac Cancer 12:1048–1057
Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G (2017) Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov 16:487–511
Mulcahy Levy JM, Thorburn A (2020) Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ 27:843–857
Saxena R, Klochkova A, Murray MG et al (2019) Roles for autophagy in esophageal carcinogenesis: implications for improving patient outcomes. Cancers (Basel) 11
Ruifrok AC, Katz RL, Johnston DA (2003) Comparison of quantification of histochemical staining by hue-saturation-intensity (HSI) transformation and color-deconvolution. App Imm Mol Morph 11:85–91
Stephen HC, Emma T, Timothy K et al (2012) Automatic nonsubjective estimation of antigen content visualized by immunohistochemistry using color deconvolution. App Imm Mol Morph 20:82–90
Nunes TWN, Filippi-Chiela EC, Callegari-Jacques SM et al (2019) Nuclear morphometric analysis in tissue as an objective tool with potential use to improve melanoma staging. Melanoma Res
Nagtegaal ID, Odze RD, Klimstra D et al (2020) The 2019 WHO classification of tumours of the digestive system. Histopathology 76:182–188
Chandrashekar DS, Bashel B, Balasubramanya SAH et al (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19:649–658
Győrffy B (2021) Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J 19:4101–4109
Tate JG, Bamford S, Jubb HC et al (2019) COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 47:D941–D947
Baek SH, Kim KI (2017) Epigenetic control of autophagy: nuclear events gain more attention. Mol Cell 65:781–785
Sui X, Zhu J, Zhou J et al (2015) Epigenetic modifications as regulatory elements of autophagy in cancer. Cancer Lett 360:106–113
Bhol CS, Panigrahi DP, Praharaj PP et al (2020) Epigenetic modifications of autophagy in cancer and cancer therapeutics. Semin Cancer Biol 66:22–33
Zink D, Fischer AH, Nickerson JA (2004) Nuclear structure in cancer cells. Nat Rev Cancer 4:677–687
Easwaran HP, Baylin SB (2010) Role of nuclear architecture in epigenetic alterations in cancer. Cold Spring Harb Symp Quant Biol 75:507–515
Fischer EG (2020) Nuclear morphology and the biology of cancer cells. Acta Cytol 64:511–519
Uhler C, Shivashankar GV (2018) Nuclear mechanopathology and cancer diagnosis. Trends Cancer 4:320–331
Wolberg WH, Street WN, Mangasarian OL (1999) Importance of nuclear morphology in breast cancer prognosis. Clin Cancer Res 5:3542–3548
Buhmeida A, Ristamäki R, Lamlum H et al (2005) Nuclear area is a prognostic determinant in advanced colorectal cancer. Anticancer Res 25:3083–3088
He Y, Zhao X, Subahan NR et al. The prognostic value of autophagy-related markers beclin-1 and microtubule-associated protein light chain 3B in cancers: a systematic review and meta-analysis. Tumour Biol 35:7317–26
Li J, Yan Q, Liu N et al (2020) The prognostic value of autophagy-related markers Bclin-1 and LC-3 in colorectal cancers: a systematic review and meta-analysis. Evid Based Complement Alternat Med 23:8475840
Zheng T, Li D, He Z et al (2018) Prognostic and clinicopathological significance of Beclin-1 in non-small-cell lung cancer: a meta-analysis. Onco Targets Ther 11:4167–4175
Ahn CH, Jeong EG, Lee JW et al (2007) Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS 115:1344–1349
Cai M, Hu Z, Liu J et al (2014) Beclin 1 expression in ovarian tissues and its effects on ovarian cancer prognosis. Int J Mol Sci 15:5292–5303
Li X, Xu H, Ma H (2013) Beclin 1 is highly expressed in papillary thyroid carcinoma and correlates with lymph node metastasis. Acta Chir Belg 113:175–181
Liu JL, Chen FF, Chang SF et al (2015) Expression of Beclin family proteins is associated with tumor progression in oral cancer. PLoS ONE 10:e0141308
Al-Shenawy HA (2016) Expression of Beclin-1, an autophagy-related marker, in chronic hepatitis and hepatocellular carcinoma and its relation with apoptotic markers. APMIS 124:229–237
Weh KM, Howell AB, Kresty LA (2016) Expression, modulation, and clinical correlates of the autophagy protein Beclin-1 in esophageal adenocarcinoma. Mol Carcinog 55:1876–1885
Shen MX, Ding JB (2017) Expression levels and roles of EMC-6, Beclin1, and Rab5a in the cervical cancer. Eur Rev Med Pharmacol Sci 21:3038–3046
Du H, Luo F, Shi M et al (2021) Beclin-1 is a promising prognostic biomarker in a specific esophageal squamous cell carcinoma population. Pathol Oncol Res 27:594724
Roesly HB, Khan MR, Chen HD et al (2012) The decreased expression of Beclin-1 correlates with progression to esophageal adenocarcinoma: the role of deoxycholic acid. Am J Physiol Gastrointest Liver Physiol 302:G864–G872
Giatromanolaki A, Koukourakis MI, Koutsopoulos A, Chloropoulou P, Liberis V, Sivridis E (2011) High Beclin 1 expression defines a poor prognosis in endometrial adenocarcinomas. Gynecol Oncol 123:147–151
Wang J, Pan XL, Ding LJ, Liu DY, Da-Peng Lei Jin T (2013) Aberrant expression of Beclin-1 and LC3 correlates with poor prognosis of human hypopharyngeal squamous cell carcinoma. PLoS One 8:e69038
Ju LL, Zhao CY, Ye KF, Yang H, Zhang J (2016) Expression and clinical implication of Beclin1, HMGB1, p62, survivin, BRCA1 and ERCC1 in epithelial ovarian tumor tissues. Eur Rev Med Pharmacol Sci 20:1993–2003
Valente G, Morani F, Nicotra G et al (2014) Expression and clinical significance of the autophagy proteins BECLIN 1 and LC3 in ovarian cancer. Biomed Res Int 2014:462658
Minamoto T, Nakayama K, Nakamura K et al (2018) Loss of beclin 1 expression in ovarian cancer: a potential biomarker for predicting unfavorable outcomes. Oncol Lett 15:1170–1176
Liu JL, Chen FF, Lung J et al (2014) Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br J Cancer 111:944–954
Sakurai T, Okumura H, Matsumoto M et al (2013) The expression of LC-3 is related to tumor suppression through angiogenesis in esophageal cancer. Med Oncol 30:701
Winardi D, Tsai HP, Chai CY et al (2014) Correlation of altered expression of the autophagy marker LC3B with poor prognosis in astrocytoma. Biomed Res Int 2014:723176
Lee YJ, Hah YJ, Ha YJ et al (2013) The autophagy-related marker LC3 can predict prognosis in human hepatocellular carcinoma. PLoS ONE 8:e81540
Inui T, Chano T, Takikita-Suzuki M, Nishikawa M, Yamamoto G, Okabe H (2013) Association of p62/SQSTM1 excess and oral carcinogenesis. PLoS ONE 8:e74398
Kuo WL, Sharifi MN, Lingen MW et al (2014) p62/SQSTM1 accumulation in squamous cell carcinoma of head and neck predicts sensitivity to phosphatidylinositol 3-kinase pathway inhibitors. PLoS ONE 9:e90171
Schläfli AM, Adams O, Galván JA et al (2016) Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget 7:39544–39555
Qian HL, Peng XX, Chen SH, Ye HM, Qiu JH (2005) p62 Expression in primary carcinomas of the digestive system. World J Gastroenterol 11:1788–1792
Shi C, Pan BQ, Shi F et al (2018) Sequestosome 1 protects esophageal squamous carcinoma cells from apoptosis via stabilizing SKP2 under serum starvation condition. Oncogene 37:3260–3274
Cui H, Weng Y, Ding N et al (2021) Autophagy-related three-gene prognostic signature for predicting survival in esophageal squamous cell carcinoma. Front Oncol 11:650891
Duan L, Cao L, Zhang R et al (2021) Development and validation of a survival model for esophageal adenocarcinoma based on autophagy-associated genes. Bioengineered 12:3434–3454
Yang C, Shen S, Zheng X et al (2020) Long non-coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. FASEB J 34:6055–6069
Bártová E, Krejcí J, Harnicarová A, Galiová G, Kozubek S (2008) Histone modifications and nuclear architecture: a review. J Histochem Cytochem 56:711–721
Veltri RW, Christudass CS (2014) Nuclear morphometry, epigenetic changes, and clinical relevance in prostate cancer. Adv Exp Med Biol 773:77–99
Miyamoto H, Isobe H, Akita H et al (1992) The flow cytometric nuclear-DNA content, tumor-origin, nuclear size and prognosis in squamous-cell lung-cancer. Int J Oncol 1:325–329
Buhmeida A, Algars A, Ristamäki R, Collan Y, Syrjänen K, Pyrhönen S (2006) Nuclear size as prognostic determinant in stage II and stage III colorectal adenocarcinoma. Anticancer Res 26:455–462
Nakazato Y, Minami Y, Kobayashi H et al (2010) Nuclear grading of primary pulmonary adenocarcinomas: correlation between nuclear size and prognosis. Cancer 116:2011–2019
Shin HR, Kim H, Kim KI, Baek SH (2016) Epigenetic and transcriptional regulation of autophagy. Autophagy 12:2248–2249
Acknowledgements
We thank the Research and Event Incentive Fund (FIPE) of the Hospital de Clínicas of Porto Alegre (HCPA) for funding this study and the statistical analysis service of the Hospital de Clínicas de Porto Alegre for helping with the project.
Funding
This study was funded by the Research and Event Incentive Fund (FIPE) of the Hospital de Clínicas of Porto Alegre (HCPA), project no. 16–0233.
Author information
Authors and Affiliations
Contributions
RI, EFSP, and ECFC contributed to the study conception and design. RI, PF, PLDCL, and SMCJ performed material preparation and data collection and analysis. FH and FV performed histopathological analysis. RI and ECFC wrote the first draft of the manuscript, and all authors commented on previous versions of the article. ADBL reviewed and tabulated patients’ medical records in the Hospital de Clínicas de Porto Alegre system and also reviewed the manuscript. All authors read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures performed in this study, from sample recruitment and use, as well as access to clinical information were approved by the Ethical Committee of the Hospital de Clinicas de Porto Alegre under the Certificate of Presentation for Ethical Appreciation (CAAE): 57102216.0.0000.5327. The risk of breach of confidentiality was controlled by signing the term of responsibility for the use of biological material, in which the researchers undertake not to use references that identify the study patients.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Iserhard, R., Pilar, E.F.S., de Oliveira, F.H. et al. Autophagy and nuclear morphometry are associated with histopathologic features in esophageal squamous cell carcinoma. J Mol Med 102, 39–52 (2024). https://doi.org/10.1007/s00109-023-02387-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-023-02387-4