Abstract
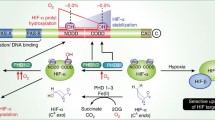
In order to pass through the nuclear pore complex, proteins larger than ∼40 kDa require specific nuclear transport receptors. Defects in nuclear-cytoplasmatic transport affect fundamental processes such as development, inflammation and oxygen sensing. The transcriptional response to O2 deficiency is controlled by hypoxia-inducible factors (HIFs). These are heterodimeric transcription factors of each ∼100–120 kDa proteins, consisting of one out of three different O2-labile α subunits (primarily HIF-1α) and a more constitutive 1β subunit. In the presence of O2, the α subunits are hydroxylated by specific prolyl-4-hydroxylase domain proteins (PHD1, PHD2, and PHD3) and an asparaginyl hydroxylase (factor inhibiting HIF-1, FIH-1). The prolyl hydroxylation causes recognition by von Hippel-Lindau tumor suppressor protein (pVHL), ubiquitination, and proteasomal degradation. The activity of the oxygen sensing machinery depends on dynamic intracellular trafficking. Nuclear import of HIF-1α and HIF-1β is mainly mediated by importins α and β (α/β). HIF-1α can shuttle between nucleus and cytoplasm, while HIF-1β is permanently inside the nucleus. pVHL is localized to both compartments. Nuclear import of PHD1 relies on a nuclear localization signal (NLS) and uses the classical import pathway involving importin α/β receptors. PHD2 shows an atypical NLS, and its nuclear import does not occur via the classical pathway. PHD2-mediated hydroxylation of HIF-1α occurs predominantly in the cell nucleus. Nuclear export of PHD2 involves a nuclear export signal (NES) in the N-terminus and depends on the export receptor chromosome region maintenance 1 (CRM1). Nuclear import of PHD3 is mediated by importin α/β receptors and depends on a non-classical NLS. Specific modification of the nuclear translocation of the three PHD isoforms could provide a promising strategy for the development of new therapeutic substances to tackle major diseases.


Similar content being viewed by others
References
Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15:607–660
Tran EJ, King MC, Corbett AH (2014) Macromolecular transport between the nucleus and the cytoplasm: advances in mechanism and emerging links to disease. Biochim Biophys Acta 1843:2784–2795
Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem 282:5101–5105
Conti E, Uy M, Leighton L, Blobel G, Kuriyan J (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193–204
Fontes MR, Teh T, Kobe B (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol 297:1183–1194
Friedrich B, Quensel C, Sommer T, Hartmann E, Kohler M (2006) Nuclear localization signal and protein context both mediate importin alpha specificity of nuclear import substrates. Mol Cell Biol 26:8697–8709
Fagerlund R, Melen K, Cao X, Julkunen I (2008) NF-kappaB p52, RelB and c-Rel are transported into the nucleus via a subset of importin alpha molecules. Cell Signal 20:1442–1451
Riddick G, Macara IG (2007) The adapter importin-alpha provides flexible control of nuclear import at the expense of efficiency. Mol Syst Biol 3:118
Kimura M, Imamoto N (2014) Biological significance of the importin-beta family-dependent nucleocytoplasmic transport pathways. Traffic 15:727–748
Flores K, Seger R (2013) Stimulated nuclear import by beta-like importins. F1000Prime Rep 5:41
Chook YM, Suel KE (2011) Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta 1813:1593–1606
Magnani M, Crinelli R, Bianchi M, Antonelli A (2000) The ubiquitin-dependent proteolytic system and other potential targets for the modulation of nuclear factor-kB (NF-kB). Curr Drug Targets 1:387–399
la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17:527–536
Kutay U, Guttinger S (2005) Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15:121–124
Hutten S, Kehlenbach RH (2007) CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 17:193–201
Semenza GL (2013) HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest 123:3664–3671
Huenniger K, Kramer A, Soom M, Chang I, Kohler M, Depping R, Kehlenbach RH, Kaether C (2010) Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cell Mol Life Sci 67:3187–3196
Jang AR, Moravcevic K, Saez L, Young MW, Sehgal A (2015) Drosophila TIM binds importin alpha1, and acts as an adapter to transport PER to the nucleus. PLoS Genet 11:e1004974
Depping R, Steinhoff A, Schindler SG, Friedrich B, Fagerlund R, Metzen E, Hartmann E, Kohler M (2008) Nuclear translocation of hypoxia-inducible factors (HIFs): Involvement of the classical importin alpha/beta pathway. Biochim Biophys Acta 1783:394–404
Suarez-Sanchez R, Aguilar A, Wagstaff KM, Velez G, Zuara-Medina PM, Gomez P, Vasquez-Limeta A, Hernandez-Hernandez O, Lieu KG, Jans DA et al (2014) Nucleocytoplasmic shuttling of the Duchenne muscular dystrophy gene product dystrophin Dp71d is dependent on the importin alpha/beta and CRM1 nuclear transporters and microtubule motor dynein. Biochim Biophys Acta 1843:985–1001
Takeda E, Murakami T, Matsuda G, Murakami H, Zako T, Maeda M, Aida Y (2011) Nuclear exportin receptor CAS regulates the NPI-1-mediated nuclear import of HIV-1 Vpr. PLoS One 6:e27815
Ao Z, Danappa JK, Wang B, Zheng Y, Kung S, Rassart E, Depping R, Kohler M, Cohen EA, Yao X (2010) Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J Virol 84:8650–8663
Panchal M, Rawat K, Kumar G, Kibria KM, Singh S, Kalamuddin M, Mohmmed A, Malhotra P, Tuteja R (2014) Plasmodium falciparum signal recognition particle components and anti-parasitic effect of ivermectin in blocking nucleo-cytoplasmic shuttling of SRP. Cell Death Dis 5:e994
Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K (2005) NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem 280:15942–15951
Kim IS, Kim DH, Han SM, Chin MU, Nam HJ, Cho HP, Choi SY, Song BJ, Kim ER, Bae YS et al (2000) Truncated form of importin alpha identified in breast cancer cell inhibits nuclear import of p53. J Biol Chem 275:23139–23145
Depping R, Schindler SG, Jacobi C, Kirschner KM, Scholz H (2012) Nuclear transport of Wilms’ tumour protein Wt1 involves importins alpha and beta. Cell Physiol Biochem 29:223–232
Nagara Y, Tateishi T, Yamasaki R, Hayashi S, Kawamura M, Kikuchi H, Iinuma KM, Tanaka M, Iwaki T, Matsushita T et al (2013) Impaired cytoplasmic-nuclear transport of hypoxia-inducible factor-1alpha in amyotrophic lateral sclerosis. Brain Pathol 23:534–546
Kose S, Imamoto N (2014) Nucleocytoplasmic transport under stress conditions and its role in HSP70 chaperone systems. Biochim Biophys Acta 1840:2953–2960
Jo S, Kallo I, Bardoczi Z, Drigo A e, Drigo A e, Zeold A, Liposits Z, Oliva A, Lemmon VP, Bixby JL et al (2012) Neuronal hypoxia induces Hsp40-mediated nuclear import of type 3 deiodinase as an adaptive mechanism to reduce cellular metabolism. J Neurosci 32:8491–8500
Crampton N, Kodiha M, Shrivastava S, Umar R, Stochaj U (2009) Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol Biol Cell 20:5106–5116
Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, Petruzziello-Pellegrini TN, Tsui AK, He JZ, Dhamko H et al (2012) Functional importance of Dicer protein in the adaptive cellular response to hypoxia. J Biol Chem 287:29003–29020
Semenza GL (2010) Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29:625–634
Mandl M, Kapeller B, Lieber R, Macfelda K (2013) Hypoxia-inducible factor-1beta (HIF-1beta) is upregulated in a HIF-1alpha-dependent manner in 518A2 human melanoma cells under hypoxic conditions. Biochem Biophys Res Commun 434:166–172
Mandl M, Depping R (2014) Hypoxia-inducible aryl hydrocarbon receptor nuclear translocator (ARNT) (HIF-1beta): is it a rare exception? Mol Med 20:215–220
Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A et al (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43–54
Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 16:1466–1471
Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY (2002) Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 417:975–978
Jiang BH, Semenza GL, Bauer C, Marti HH (1996) Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol 271:C1172–C1180
Wang GL, Semenza GL (1995) Purification and characterization of hypoxia-inducible factor 1. J Biol Chem 270:1230–1237
Kallio PJ, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L (1998) Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J 17:6573–6586
Luo JC, Shibuya M (2001) A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1alpha, 2alpha and 3alpha). Oncogene 20:1435–1444
Chachami G, Paraskeva E, Mingot JM, Braliou GG, Gorlich D, Simos G (2009) Transport of hypoxia-inducible factor HIF-1alpha into the nucleus involves importins 4 and 7. Biochem Biophys Res Commun 390:235–240
Mylonis I, Chachami G, Paraskeva E, Simos G (2008) Atypical CRM1-dependent nuclear export signal mediates regulation of hypoxia-inducible factor-1alpha by MAPK. J Biol Chem 283:27620–27627
Tian H, McKnight SL, Russell DW (1997) Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev 11:72–82
Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii KY (1997) A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U S A 94:4273–4278
Flamme I, Frohlich T, von Reutern M, Kappel A, Damert A, Risau W (1997) HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1α and developmentally expressed in blood vessels. Mech Dev 63:51–60
O'Rourke JF, Tian YM, Ratcliffe PJ, Pugh CW (1999) Oxygen-regulated and transactivating domains in endothelial PAS protein 1: comparison with hypoxia-inducible factor-1alpha. J Biol Chem 274:2060–2071
Zhao J, Du F, Shen G, Zheng F, Xu B (2015) The role of hypoxia-inducible factor-2 in digestive system cancers. Cell Death Dis 6:e1600
Keith B, Johnson RS, Simon MC (2011) HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12:9–22
Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC (2003) Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol 23:9361–9374
Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, Eckardt KU (2004) Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J 18:1462–1464
Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L (2002) Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3α locus. J Biol Chem 277:32405–32408
Heikkila M, Pasanen A, Kivirikko KI, Myllyharju J (2011) Roles of the human hypoxia-inducible factor (HIF)-3alpha variants in the hypoxia response. Cell Mol Life Sci 68:3885–3901
Yamashita T, Ohneda O, Nagano M, Iemitsu M, Makino Y, Tanaka H, Miyauchi T, Goto K, Ohneda K, Fujii-Kuriyama Y et al (2008) Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol 28:1285–1297
Hara S, Hamada J, Kobayashi C, Kondo Y, Imura N (2001) Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: suppression of HIF-mediated gene expression by HIF-3alpha. Biochem Biophys Res Commun 287:808–813
Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L (2001) Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 414:550–554
Maynard MA, Evans AJ, Hosomi T, Hara S, Jewett MA, Ohh M (2005) Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J 19:1396–1406
Torii S, Goto Y, Ishizawa T, Hoshi H, Goryo K, Yasumoto K, Fukumura H, Sogawa K (2011) Pro-apoptotic activity of inhibitory PAS domain protein (IPAS), a negative regulator of HIF-1, through binding to pro-survival Bcl-2 family proteins. Cell Death Differ 18:1711–1725
Myllyharju J (2013) Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol (Oxf) 208:148–165
Katschinski DM (2009) In vivo functions of the prolyl-4-hydroxylase domain oxygen sensors: direct route to the treatment of anaemia and the protection of ischaemic tissues. Acta Physiol (Oxf) 195:407–414
Oehme F, Ellinghaus P, Kolkhof P, Smith TJ, Ramakrishnan S, Hütter J, Schramm M, Flamme I (2002) Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors. Biochem Biophys Res Commun 296:343–349
Cioffi CL, Liu XQ, Kosinski PA, Garay M, Bowen BR (2003) Differential regulation of HIF-1α prolyl-4-hydroxylase genes by hypoxia in human cardiovascular cells. Biochem Biophys Res Commun 303:947–953
Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM (2004) Differential function of the prolyl hydroxylases, PHD1, 2 and 3 in the regulation of hypoxia inducible factor (HIF). J Biol Chem 279:38458–38465
Koivunen P, Tiainen P, Hyvarinen J, Williams KE, Sormunen R, Klaus SJ, Kivirikko KI, Myllyharju J (2007) An endoplasmic reticulum transmembrane prolyl 4-hydroxylase is induced by hypoxia and acts on hypoxia-inducible factor alpha. J Biol Chem 282:30544–30552
Mylonis I, Chachami G, Samiotaki M, Panayotou G, Paraskeva E, Kalousi A, Georgatsou E, Bonanou S, Simos G (2006) Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor 1α. J Biol Chem 281:33095–33106
Carbonaro M, Escuin D, O'Brate A, Thadani-Mulero M, Giannakakou P (2012) Microtubules regulate hypoxia-inducible factor-1alpha protein trafficking and activity: implications for taxane therapy. J Biol Chem 287:11859–11869
Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A (1996) Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci 21:59–64
Ahluwalia A, Narula J, Jones MK, Deng X, Tarnawski AS (2010) Impaired angiogenesis in aging myocardial microvascular endothelial cells is associated with reduced importin alpha and decreased nuclear transport of HIF1 alpha: mechanistic implications. J Physiol Pharmacol 61:133–139
Nardozzi JD, Lott K, Cingolani G (2010) Phosphorylation meets nuclear import: a review. Cell Commun Signal 8:32
Pichler A, Melchior F (2002) Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381–387
Golan M, Mabjeesh NJ (2013) SEPT9_i1 is required for the association between HIF-1alpha and importin-alpha to promote efficient nuclear translocation. Cell Cycle 12:2297–2308
Torii S, Sakaki K, Otomo M, Saka K, Yasumoto K, Sogawa K (2013) Nucleocytoplasmic shuttling of IPAS by its unique nuclear import and export signals unshared with other HIF-3alpha splice variants. J Biochem 154:561–567
Eguchi H, Ikuta T, Tachibana T, Yoneda Y, Kawajiri K (1997) A nuclear localization signal of human aryl hydrocarbon receptor nuclear translocator/hypoxia-inducible factor 1beta is a novel bipartite type recognized by the two components of nuclear pore-targeting complex. J Biol Chem 272:17640–17647
Wang GL, Jiang BH, Rue EA, Semenza GL (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92:5510–5514
Kallio PJ, Pongratz I, Gradin K, McGuire J, Poellinger L (1997) Activation of hypoxia-inducible factor 1alpha: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc Natl Acad Sci U S A 94:5667–5672
Mandl M, Depping R (2014) Hypoxia-inducible ARNT (HIF-1beta): a rare exception? Mol Med 10
Metzen E, Berchner-Pfannschmidt U, Stengel P, Marxsen JH, Stolze I, Klinger M, Huang WQ, Wotzlaw C, Hellwig-Buergel T, Jelkmann W et al (2003) Intracellular localisation of human HIF-1α hydroxylases: implications for oxygen sensing. J Cell Sci 116:1319–1326
Steinhoff A, Pientka FK, Mockel S, Kettelhake A, Hartmann E, Kohler M, Depping R (2009) Cellular oxygen sensing: Importins and exportins are mediators of intracellular localisation of prolyl-4-hydroxylases PHD1 and PHD2. Biochem Biophys Res Commun 387:705–711
Wotzlaw C, Gneuss S, Konietzny R, Fandrey J (2010) Nanoscopy of the cellular response to hypoxia by means of fluorescence resonance energy transfer (FRET) and new FRET software. PMC Biophys 3:5
Yasumoto K, Kowata Y, Yoshida A, Torii S, Sogawa K (2009) Role of the intracellular localization of HIF-prolyl hydroxylases. Biochim Biophys Acta 1793:792–797
Berchner-Pfannschmidt U, Tug S, Trinidad B, Oehme F, Yamac H, Wotzlaw C, Flamme I, Fandrey J (2008) Nuclear oxygen sensing: induction of endogenous prolyl-hydroxylase 2 activity by hypoxia and nitric oxide. J Biol Chem 283:31745–31753
Berra E, Roux D, Richard DE, Pouyssegur J (2001) Hypoxia-inducible factor-1α (HIF-1α) escapes O2-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO J 2:615–620
Jokilehto T, Rantanen K, Luukkaa M, Heikkinen P, Grenman R, Minn H, Kronqvist P, Jaakkola PM (2006) Overexpression and nuclear translocation of hypoxia-inducible factor prolyl hydroxylase PHD2 in head and neck squamous cell carcinoma is associated with tumor aggressiveness. Clin Cancer Res 12:1080–1087
Soilleux EJ, Turley H, Tian YM, Pugh CW, Gatter KC, Harris AL (2005) Use of novel monoclonal antibodies to determine the expression and distribution of the hypoxia regulatory factors PHD-1, PHD-2, PHD-3 and FIH in normal and neoplastic human tissues. Histopathology 47:602–610
Pientka FK, Hu J, Schindler S, Brix B, Johren O, Fandrey J, Berchner-Pfannschmidt U, Depping R (2012) Oxygen sensing by prolyl-4-hydroxylase PHD2 within the nuclear compartment and the influence of compartimentalization on HIF-1 signaling. J Cell Sci 125:5168–5176
McDonough MA, Li V, Flashman E, Chowdhury R, Mohr C, Lienard BM, Zondlo J, Oldham NJ, Clifton IJ, Lewis J et al (2006) Cellular oxygen sensing: crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc Natl Acad Sci U S A 103:9814–9819
Groulx I, Lee S (2002) Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol 22:5319–5336
Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH (2006) Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol 26:8336–8346
Takeda K, Cowan A, Fong GH (2007) Essential role for prolyl hydroxylase domain protein 2 in oxygen homeostasis of the adult vascular system. Circulation 116:774–781
Takeda K, Aguila HL, Parikh NS, Li X, Lamothe K, Duan LJ, Takeda H, Lee FS, Fong GH (2008) Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood 111:3229–3235
Shin D, Jeon JH, Jeong M, Suh HW, Kim S, Kim HC, Moon OS, Kim YS, Chung JW, Yoon SR et al (2008) VDUP1 mediates nuclear export of HIF1alpha via CRM1-dependent pathway. Biochim Biophys Acta 1783:838–848
Huang J, Zhao Q, Mooney SM, Lee FS (2002) Sequence determinants in hypoxia inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J Biol Chem 277:39792–39800
Hilz A, Schillinger T, Schindler S, Köster M, Depping R (2007) Characterisation of NLS in human PHD1 and PHD3. FEBS J 274
Linke S, Stojkoski C, Kewley RJ, Booker GW, Whitelaw ML, Peet DJ (2004) Substrate requirements of the oxygen-sensing asparaginyl hydroxylase factor-inhibiting hypoxia-inducible factor. J Biol Chem 279:14391–14397
Groulx I, Lee S (2002) Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol 22:5319–5336
Khacho M, Mekhail K, Pilon-Larose K, Payette J, Lee S (2008) Cancer-causing mutations in a novel transcription-dependent nuclear export motif of VHL abrogate oxygen-dependent degradation of hypoxia-inducible factor. Mol Cell Biol 28:302–314
Khacho M, Lee S (2009) Subcellular dynamics of the VHL tumor suppressor: on the move for HIF degradation. Future Oncol 5:85–95
Cai Q, Robertson ES (2010) Ubiquitin/SUMO modification regulates VHL protein stability and nucleocytoplasmic localization. PLoS One 5:e12636
Jokilehto T, Hogel H, Heikkinen P, Rantanen K, Elenius K, Sundstrom J, Jaakkola PM (2010) Retention of prolyl hydroxylase PHD2 in the cytoplasm prevents PHD2-induced anchorage-independent carcinoma cell growth. Exp Cell Res 316:1169–1178
Jokilehto T, Jaakkola PM (2010) The role of HIF prolyl hydroxylases in tumour growth. J Cell Mol Med 14:758–770
Luukkaa M, Jokilehto T, Kronqvist P, Vahlberg T, Grenman R, Jaakkola P, Minn H (2009) Expression of the cellular oxygen sensor PHD2 (EGLN-1) predicts radiation sensitivity in squamous cell cancer of the head and neck. Int J Radiat Biol 85:900–908
Couvelard A, Deschamps L, Rebours V, Sauvanet A, Gatter K, Pezzella F, Ruszniewski P, Bedossa P (2008) Overexpression of the oxygen sensors PHD-1, PHD-2, PHD-3, and FIH Is associated with tumor aggressiveness in pancreatic endocrine tumors. Clin Cancer Res 14:6634–6639
Chan DA, Sutphin PD, Yen SE, Giaccia AJ (2005) Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1 alpha. Mol Cell Biol 25:6415–6426
Ozer A, Wu LC, Bruick RK (2005) The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF). Proc Natl Acad Sci U S A 102:7481–7486
Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, Walker A, Klisovic R, Blum W, Caligiuri M et al (2012) Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 120:1765–1773
Mallavia B, Recio C, Oguiza A, Ortiz-Munoz G, Lazaro I, Lopez-Parra V, Lopez-Franco O, Schindler S, Depping R, Egido J et al (2013) Peptide inhibitor of NF-kappaB translocation ameliorates experimental atherosclerosis. Am J Pathol 182:1910–1921
Acknowledgments
The authors gratefully acknowledge financial support by the “Werner and Klara Kreitz-Stiftung” and the “Sektion Medizin an der Universität zu Lübeck J19-2015”. The authors wish to thank G. Fletschinger for preparing the artwork.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Depping, R., Jelkmann, W. & Kosyna, F.K. Nuclear-cytoplasmatic shuttling of proteins in control of cellular oxygen sensing. J Mol Med 93, 599–608 (2015). https://doi.org/10.1007/s00109-015-1276-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-015-1276-0