Abstract
Foraging ants face many dangers in search of food and often need to defend their prey to ensure the colony’s survival, although ants may also follow a peaceful foraging strategy. A non-aggressive approach is seen in the Australian bull ant Myrmecia nigriceps, in that workers of neighboring nests sometimes share foraging trees. In this study, we observed 31 nests at Mount Majura Nature Reserve in Canberra (Australia), 12 of which shared a foraging tree with at least one other nest in at least one of three nights. We genotyped 360 individuals at five published microsatellite loci and further established a set of nine polymorphic loci for M. nigriceps. Our results revealed a significant correlation between tree sharing and geographical distance between nests. We found no correlation between internest relatedness and tree sharing, geographical distance between nests and internest relatedness, and intranest relatedness and tree sharing. We further investigated the colony structure of M. nigriceps. All colonies were monodomous; the number of queens per colony ranged from one to two, and the number of fathers from one to three. No instances of worker drifting were found in this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foraging is one of the most vital parts of survival in the life of an ant colony. However, it is also one of the costliest activities as workers spend great amounts of energy and face many risks in search of food (Dornhaus and Powell 2009). Different species of ants employ different strategies for the discovery, retrieval, and defense of food. For example, workers may forage individually or in groups and use morphological, chemical, and behavioral defenses to ensure survival and safety of resources (Hölldobler and Wilson 1990; Dornhaus and Powell 2009). As an alternative to a defensive approach, ants may also avoid aggressive behavior or share food sources (d’Ettorre and Lenoir 2009).
The ant genus Myrmecia consists of 93 described species (Hasegawa and Crozier 2006; Taylor 2015) which are endemic to Australia or, in one instance, New Caledonia. Although rejected as among the most basal formicids, ants of this genus have retained many biological traits that are considered to be archaic (Ogata and Taylor 1991; Ward and Brady 2003; Hasegawa and Crozier 2006). Commonly known as “bulldog ants” or “bull ants”, these ants are characterized by relatively large bodies, little morphological difference between queens and workers, strongly toothed, long mandibles, and aggressive behavior, which is accompanied by a painful stinger (Eriksson 1985; Ogata and Taylor 1991; Ward and Brady 2003). They forage individually, have unusually large eyes and rely extensively on vision for navigation and capture of prey. Several species operate in discrete temporal niches, ranging from strictly diurnal to crepuscular to strictly nocturnal. During these species-specific timespans, workers leave the nest individually to forage on near-standing trees (Narendra et al. 2017). On the trees, the ants hunt for a wide variety of arthropods, feed on sap produced by trees, and tend to aphids, coccids and mealybugs for honeydew (Reid et al. 2013). Evolutionary adaptations of the visual system of species to the specific light environments in which they operate show how well these ants are adapted to their temporal niches (Narendra et al. 2011, 2016, 2017). Their visual navigation and foraging ecology set bull ants up as attractive model organisms in ecological neurobiology (Narendra et al. 2016; Kamhi et al. 2020).
One such study system is Myrmecia nigriceps Mayr, 1862, which represents a crepuscular–nocturnal foraging schedule (Narendra et al. 2016). It belongs to the gulosa species group, one of nine recognized species groups within the genus Myrmecia (Ogata 1991; Ogata and Taylor 1991). Studies have focused on the species’ brains and their visual systems (Greiner et al. 2007; Narendra et al. 2011, 2016; Narendra and Ribi 2017; Sheehan et al. 2019), and little is known about its general biology, including its social structure, which is a key aspect of ant ecology. Attributes influencing a colony’s social structure include the number of queens (monogyny versus polygyny), number of fathers (monandry versus polyandry), and number of nests (monodomy versus polydomy; Steiner et al. 2009). Myrmecia nigriceps can be found in underground earth nests, indicated by characteristic gravel mounds surrounding the nest entrance. The colony comprises a few hundred workers (Shattuck 1999; van Wilgenburg et al. 2007) and can be found in habitats where colonies have access to lone standing Eucalyptus trees to forage on, such as tall grassy woodland and dry sclerophyll open forest (Baines et al. 2013). Even though workers of M. nigriceps are extremely aggressive towards hetero-specific intruders, van Wilgenburg et al. (2007) found a general absence of aggression towards conspecific nest-intruders in a series of bioassays introducing non-nestmates to foreign colonies of M. nigriceps. This peaceful behavior can sometimes also be seen when ants of adjoining colonies encounter each other while foraging. In juxtaposition, Readhead (2014), who investigated aggression and cuticular hydrocarbon profiles in a subpopulation of M. nigriceps also used in this work, found no absence of aggression but rather noted instances of avoidance.
Here, we aim to investigate the social structure of one population of M. nigriceps which has been subject to previous neurobiological studies. We try to find out whether (i) geographical distance between nests correlates with sharing of foraging trees, (ii) internest relatedness correlates with sharing of foraging trees, and (iii) geographical distance between nests correlates with internest relatedness. Further, we test for correlation of (iv) intranest relatedness with sharing of foraging trees and lastly investigate whether nests that share foraging trees represent separate colonies or if they rather belong to the same polydomous colony.
Materials and methods
Field work and DNA extraction
Field work was conducted in Canberra, Australia, from 6–19 November 2017 and 11–17 December 2017. A total of 36 nests of Myrmecia nigriceps, from three main locations, (i) Mount Majura Nature Reserve (31 nests; henceforth Majura North), (ii) a second site at Mount Majura Nature Reserve (two nests; Majura South), and (iii) the Australian National University campus (three nests; ANU; Fig. 1ab), were marked and recorded by GPS (Garmin, eTrex 10, Olathe, Kansas). We used ants from all the nests to investigate colony and population structure. Only at Majura North did we identify the trees on which ants from each nest forage. Foraging behavior was observed for three nights per nest (partially split between the two field work periods) by following individual ants from the nest entrance to their respective foraging trees, which were defined as such by workers ascending the stem for 0.5 m. To keep track of the workers without disturbing them, a headlamp with red light was used. As foraging onset regularly occurs during evening twilight and does not exhibit seasonal changes, reflected in the activity patterns of the related species Myrmecia pyriformis (Narendra et al. 2010, 2011), all observations started 30 min before sunset and ended when no more workers could be seen foraging. After completion of all foraging observations, workers were collected and stored in absolute ethanol p.a. Genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen, Hilden; Germany) according to the manufacturer’s instructions, except that proteinase K digestion was prolonged to overnight incubation.
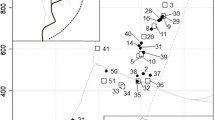
a Overview of 31 nests of Myrmecia nigriceps at Mount Majura Nature Reserve and outer nests (at Mount Majura Nature Reserve and the Australian National University campus) found in Canberra, Australia. b Close-up of nests located at Mount Majura, where foraging behavior was observed for three nights per nest. Satellite imageries were provided by nearmap (nearmap.com.au)
Primer characterization and microsatellite genotyping
Five published microsatellite loci (Mbre11(HEX), Mbre16 (FAM), Mbre17 (PET), Mbre67 (NED), Nmac18 (PET)) were selected from a set of 16 loci (Qian et al. 2011a) after testing on 24 individuals for amplification success, scorability, and allele count. A total of 360 workers, 10 ants per nest, were genotyped at these five loci. Additionally, a set of nine species-specific loci was newly developed for Myrmecia nigriceps (Table 1). For this, genomic DNA of two degastered individuals was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). DNA was quantified with ribogreen (Thermo Fisher, Waltham, USA) and tested for human contamination by AluJ real-time PCR (Kaneko et al. 2011) in triplicates; human positive controls were run as standard in a dilution series 1:1, 1:10, and 1:100. Illumina PE250 libraries were prepared by a commercial sequencing facility (IGA, Udine, Italy), and sequenced on the Illumina NextSeq 500 system. Low-quality reads were discarded and SciRoKo (Kofler et al. 2007) was used to identify di- and trinucleotide repeat units. To obtain reads with at least eight microsatellite repeat units, a 100 bp flanking region at both sides, and a balanced GC content in the flanking region, custom Python scripts were used. Microsatellite primers were designed using FastPCR v6.6.71 (Kalendar et al. 2017) and Primer3web v4.1 (Untergasser et al. 2012). All 14 loci (five published and nine novel ones mentioned above) were labeled with a fluorescent dye via M13-tailed PCR (Boutin-Ganache et al. 2001), using the following thermal profile: 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 45 s, followed by a final extension at 72 °C for 10 min. Amplifications were carried out in a 5 μl reaction volume containing 1 × Rotorgene Probe-PCR Kit Mastermix (Qiagen, Hilden, Germany), 0.02 μM forward primer, 0.2 μM M13-tailed primer, 0.2 μM reverse primer, and 0.5 μl DNA extract on a UnoCycler 1200 (VWR, Radnor, USA). Amplification success was checked by agarose gel electrophoresis. Capillary electrophoresis was performed using an ABI 3130 instrument (Applied Biosystems, Foster City, USA) at a commercial sequencing facility (CRC Sequencing Facility, Chicago, USA). Alleles were scored manually using PeakScanner v1.0 (Applied Biosystems, Foster City, USA).
Population genetic analysis
Linkage Disequilibrium (LD) was calculated using GenePop on the Web (Raymond and Rousset 1995). Deviations from Hardy–Weinberg equilibrium (HWE), F-statistics, and pairwise relatedness following Queller and Goodnight (1989) were calculated using GenAlEx v6.41 (Peakall and Smouse 2012). The mean relatedness within nests (intranest relatedness) and the mean relatedness between nests (internest relatedness) were calculated based on pairwise relatedness in GenAlEx v6.41. The possible presence of null alleles was tested using MicroChecker v2.2.3 (van Oosterhout et al. 2004). Both HWE and LD were tested on a subset of the original dataset using single worker genotypes randomly sampled from each nest. Bonferroni–Holm corrections for multiple comparisons were performed at significance level 0.05 (Armstrong 2014). After adjusting coordinates with help of orthophotos from 2014 (source: services.ga.gov.au, retrieved 26 Jun 2016) and noted position of trees, geographic distance between nests was calculated using ArcGIS v10.5.1 (www.esri.com).
Logistic regressions of (i) tree sharing against geographical distance and (ii) tree sharing against internest relatedness were performed in R v4.0.2 (R Core Team 2020) using RStudio v1.4.1103 (RStudio Team 2020). For this, only nest pairs with the potential for tree sharing were included in the analyses, meaning there was at least one tree visited by the workers of at least one nest during at least one night within a radius of 20 m for both nests; tree sharing was defined as the number of trees shared per night, averaged over three nights. A Mantel test for (iii) correlation between geographical distance and internest relatedness was performed. For the Mantel test, the function “mantel.rtest” in the package “ade4” was applied using 4999 permutations (Dray and Dufour 2007). Additionally, (iv) a logistic regression of tree sharing index TS, defined as the number of trees shared with at least one other nest out of the number of trees visited per night, averaged over three nights, against intranest relatedness was also performed using R. All logistic regressions were fit using a general linear model (GLM) with binomial error structure using the function “glm” in the R base package. Model fits were checked using the package DHARMa (Hartig 2020). All regression and Mantel test plots were created using the packages ggplot (Wickham 2016) and ggpubr (Kassambara 2020).
Parentage and sibship alignments
Parentage and sibship alignments (v) for all nests were estimated using COLONY v2.0.6.5 (Jones and Wang 2010). COLONY runs were performed using full likelihood (FL) Method, high precision, and long runtime. A test set of runs with both allelic dropout rates and other possible errors set to 0.0001, 0.0005, 0.001, 0.005, 0.01, and 0.05 was used to determine suitable error rates. For this, the error rates suggested by the program as well as improbable sibship relations (e.g., shared mothers between nests from Majura North, Majura South, and ANU) were taken into consideration. The final error rates were set to 0.0005 for all loci except Mnig 52, which was set to 0.05 and 0.0025 for allelic dropout rates and other errors, respectively. Additionally, individuals with many missing data were excluded from runs to correct for patterns created by missing values. We performed five parallel runs with different randomized seed number using the above-mentioned error rates to assure reproducibility. Assignment probability had to be at least 0.95 for all individuals of a nest for its results to be interpreted.
Results
Nest sites and foraging observations
We found an average distance of 159.90 m ± 111.88 m (mean ± standard deviation; n = 31, min 3.12 m, max 477.84 m; Fig. 1ab) between nests at Majura North. Of these 31 nests, 12 were observed sharing a foraging tree with at least one other nest in at least one of three nights (Supplement 1, Fig. 2). We observed that tree-sharing frequencies between nests ranged from 0.00 (no sharing) to 0.33 (one of three nights), 0.67 (two of three nights), and 1.00 (all three nights). We found that average tree sharing (TS) was 0.31 ± 0.43 (min 0.00, max 1.00; Supplement 1).
Basic population genetics and relatedness
We detected linkage between microsatellite loci Mnig49 and Mnig66 and deviations from HWE and the presence of null alleles at loci Mbre11, Mbre16, Mbre67, and Mnig48, leading us to exclude the latter five loci. For the remaining nine loci, no linkage and no deviation from HWE were observed; they yielded a total of 106 alleles, with an average of 11.8 alleles per locus ± 5.7 (mean ± standard deviation; min 6, max 23). We found that the average expected heterozygosity HE for each locus was 0.55 ± 0.15 (min 0.29, max 0.74). Additionally, we found that the average internest relatedness was − 0.01 ± 0.09 (min − 0.24, max 0.32), while the average intranest relatedness was 0.39 ± 0.15 (min 0.17, max 0.73; Supplement 1).
Mantel test and regressions
We found that as the geographical distance between nests decreased, tree sharing significantly increased (GLM with binomial error structure, z = − 2.089, p = 0.037, residual deviance 15.41 on 28 degrees of freedom (df), Fig. 3a). We found no effect between tree sharing and internest relatedness (GLM with binomial error structure, z = 1.367, p = 0.172, residual deviance 19.65 on 28 df, Fig. 3b), nor between geographical distance between nests and internest relatedness (Mantel test with 4999 permutations, r = − 0.20, p = 0.998, Fig. 3c). Lastly, we found no correlation between tree-sharing index TS and intranest relatedness (GLM with binomial error structure, z = 0.688, p = 0.491, residual deviance 29.82 on 24 df, Fig. 3d).
Regression and Mantel test plots. a Regression between geographical distance between nests and tree sharing (for definition, see “Materials and methods”). b Regression between internest relatedness and tree sharing. c Graphical depiction of the Mantel test between the geographical distance (log10) between nests and the internest relatedness (using pairwise distances). d Regression between intranest relatedness and tree sharing index TS (for definition, see “Materials and methods”) for each nest. All regressions were fit using a general linear model with a binomial error structure
Parentage and sibship determinations
We produced identical results for four out of five runs (COLONY), with the fifth deviating in three out of 14 nests (which remained from the original 36 nests after failing to meet the required probability). We identified no worker that shared a parent with a worker from another nest, that is, all nests sampled represented separate colonies (Supplement 2). We were able to identify both monogynous and oligogynous colonies, as well as monandrous and polyandrous colonies (Supplement 2). Out of 31 colonies at Majura North one was monogynous-monandrous. We found an average number of 1.10 ± 0.32 (min 1, max 2) queens per colony and an average number of 2.20 ± 0.63 (min 1, max 3) fathers. We detected all fullsibs (sharing both mother and father) as well as all halfsibs (sharing either mother or father) within their respective colonies.
Discussion
Tree sharing
Of 31 nests of Myrmecia nigriceps at Majura North, 12 were observed to share a foraging tree with at least one other nest. However, no relation between foraging and genetic relatedness and no worker drifting were found. Van Wilgenburg et al. (2007) tested worker behavior of M. nigriceps towards non-nestmate intruders and found that workers generally respond passively towards non-nestmate conspecifics that approach the nest entrance and that some intruders even remain in the nest for several minutes before re-emerging. In a different population of M. nigriceps, it was clear that non-nestmates tend to respond aggressively to each other and that aggressiveness increases with chemical distance (Readhead 2014), which suggests that M. nigriceps uses cuticular hydrocarbons for nestmate recognition and aligns with the fact that related Myrmecia species effectively do so as well (Dietemann et al. 2002). The correlation between tree sharing and geographical distance could purely reflect convenience for foraging, meaning that neighboring trees are more easily accessible. Possibly, though, it could also indicate that nests in close proximity may avoid aggression if necessary. It could mean a neutral situation, as some nests shared foraging trees in all nights although other trees without conspecifics from other nests were available, but tree sharing was not obligatory between all nests. As only a few individuals of a colony are tasked with foraging, ants can benefit by sharing resources and thus avoiding the high costs of aggression (Ellis and Robinson 2015). This might be even more relevant for M. nigriceps, where all individuals of a colony must forage. In a colony as small as that of M. nigriceps, the loss of individual workers might be nontrivial, which could explain non-aggressiveness between non-nestmates (van Wilgenburg et al. 2007). In this case, specifically, it could mean that foragers employ avoidance behavior as a strategy to reduce the cost of fighting as observed by Readhead (2014). Furthermore, the small nest size and the corresponding number of foragers might be low enough for the ants not to encounter one another on the trees. In the trap-jawed ant Daceton armigerum, which also forages singly and hunts on sight, sharing of trees (and even foraging trails) was also observed with colonies of other arboreal ants; those encounters are not always peaceful as D. armigerum often kills these other ants and steals their prey (Dejean et al. 2012). As no such behavior is recorded for M. nigriceps, it seems that if trees offer a great amount of space, food resources are not limiting within a particular foraging tree for these ants (Reid et al. 2013).
Colony structure
We found that all colonies were monodomous. The average intracolony relatedness of Myrmecia nigriceps was 0.39 ± 0.15, the number of queens ranged from one to two per colony, and the number of fathers from two to three. Generally, our findings match those of studies performed with species closely related with M. nigriceps. When comparing the results in detail, intracolony relatedness was higher in Myrmecia pyriformis (0.51 ± 0.04; Sanetra 2011), lower in Myrmecia brevinoda (0.29 ± 0.01; Qian et al. 2011c), and lower in Myrmecia pilosula (0.31 ± 0.01; Qian et al. 2011b) than that in M. nigriceps. The first evidence of facultative polygyny in the Myrmecia gulosa group was found by Dietemann et al. (2002), who identified two queens in Myrmecia gulosa in two instances. Facultative polygyny was also found in M. pilosula, where one to four queens were identified, in M. brevinoda, where one to six queens were found, and in M. pavida, where one to five queens were found (Qian et al. 2011b, c; Chappell et al. 2013). Findings for M. pyriformis by Sanetra (2011) also match ours, in that the number of queens ranged from one to two per colony in that species. Facultative polyandry was found in some of the bull ant species mentioned above, with one to two fathers in M. pilosula, one to ten in M. brevinoda, one to seven in M. pavida, and one to four in M. pyriformis (Qian et al. 2011b, c; Sanetra 2011; Chappell et al. 2013).
Polyandry vs. polygyny
Both polygyny and polyandry can lead to an increase in intracolonial genetic variability, but due to costs alongside the benefits thereof, Kellner and Reeve (2014) argued that polygyny and polyandry should be negatively associated. This hypothesis has been subject to discussion as evidence for this debated hypothesis was lacking for long at the intraspecific level (Qian et al. 2011c). In 2011, Qian et al. (2011c) found empirical intraspecific support for M. brevinoda, a species belonging to the gulosa group along M. nigriceps. While it is not possible to make a strong statement about M. nigriceps regarding polyandry vs. polygyny as there were only a few nests where assignment of parental IDs was significant, the available data suggest a tendency for M. nigriceps to follow polyandry. In polygynous colonies, queens were more likely to mate with fewer males (one to two) rather than three or more. This, alongside with similar results for the other species discussed previously, would suggest support for the polyandry vs. polygyny-hypothesis.
Population structure and dispersal
In most ant species, males and queens perform a nuptial flight to mate and found a new colony, although flightless reproductive castes do occur in some species (Helms 2017). Ants of the genus Myrmecia mate on hilltops, where they form large balls in which numerous males try to copulate with a female (Wheeler 1916). While young queens of monogynous species usually disperse to found their colony either alone or with other young queens (independent colony foundation, ICF), those of polygynous species often mate in or near their maternal colonies and often need help from workers of their natal colonies to form a new colony (dependent colony foundation, DCF) (Qian et al. 2011c). Ant queens generally vary in body size, wing size, and other aspects of flight ecology, leading to a reflection thereof in their respective dispersal distances (Helms 2017). A polymorphism of queens and differences in mating strategies in Tetramorium moravicum, where microgyny-polygyny (several small queens, mating inside the nest) exists alongside macrogyny-monogyny (large single queens, mating outside the nest) shows an example of different dispersal even within one species (Schlick-Steiner et al. 2007). Not only are the differences in mating strategies often reflected in ant morphology, indication should also be found regarding intercolony relatedness and geographical distance between colonies. Here, we found no correlation between intercolony relatedness and geographical distance (Mantel test). Mantel tests are heavily discussed among researchers and often criticized for their increased type 1 error (Diniz-Filho et al. 2013; Guillot and Rousset 2013; Crabot et al. 2018), but we note that this critique does not apply here as the correlation between relatedness and geographical distance was not significant. Although ICF and DCF may co-occur in facultative polygynous colonies, the lack of correlation between intercolony relatedness and geographical distance suggests a prevalence of ICF in M. nigriceps (Qian et al. 2011c). The graphical illustration of the Mantel test rather corresponds to a mixed strategy of short- and long-distance dispersal in these ants than to what would be expected under an exclusively short- or long-distance dispersal (Fig. 3c, Supplement 3 and 4). This seems plausible when considering the hilltop mating of M. nigriceps, which takes place after flying off from the vicinity of the colony (Wheeler 1916; Narendra et al. 2011).
Conclusion and outlook
In conclusion, we report a negative relation between tree sharing and geographical distance, as well as monogyny and oligogyny, and monandry and polyandry for Myrmecia nigriceps at Mount Majura Nature Reserve, Canberra, Australia. To further interpret the results presented in this work, an investigation of cuticular hydrocarbons and behavioral assays (especially using ants from non-neighboring colonies), as implemented by Krapf et al. (2018), could be useful, as it might help to classify their behavior in more detail and provide insight in whether or not nestmate recognition is intact in these ants; ideally, also a potential effect of seasonality on nestmate recognition should be assessed (cf. Katzerke et al. 2006; Thurin and Aron 2008). All this would help to better understand their tendency to be less aggressive when encountering foragers of another colony. Lastly, an analysis of diet could provide insight in whether different colonies have specialized on different food sources, which might explain the non-aggressive behavior when non-nestmates encounter each other on foraging trees due to non-overlapping trophic niches.
Availability of data and material
Available from the corresponding author upon request.
Code availability
Not applicable.
References
Armstrong RA (2014) When to use the Bonferroni correction. Ophthalmic Physiol Opt 34:502–508
Baines G, Webster M, Cook E, Johnston L, Seddon J (2013) The vegetation of the Kowen, Majura and Jerrabomberra districts of the Australian Capital Territory. Conservation Planning and Research, ACT Government, pp 21–25
Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques 31:24–26
Chappell P, Roberts K, Baer B, Hughes WHO (2013) Mating system and population genetic structure of the bulldog ant Myrmecia pavida (Hymenoptera: Formicidae). Myrmecol News 18:25–32
Crabot J, Clappe S, Dray S, Datry T (2018) Testing the Mantel statistic with a spatially-constrained permutation procedure. Methods Ecol Evol 10:532–540
d’Ettorre P, Lenoir A (2009) Nestmate recognition. In: Lach L, Parr CL, Abbott KL (eds) Ant ecology. Oxford University Press, Oxford, pp 194–209
Dejean A, Delabie JHC, Corbara B, Azemar F, Groc S, Orivel J, Leponce M (2012) The ecology and feeding habits of the arboreal trap-jawed ant Daceton armigerum. PLoS ONE 7:e37683
Dietemann V, Hölldobler B, Peeters C (2002) Caste specialisation and differentation in reproductive potential in the phylogenetically primitive ant Myrmecia gulosa. Insectes Soc 49:289–298
Diniz-Filho JAF, Soares TH, Lima JS, Dobrovolski R, Landeiro VL, de Campos Telles MP, Rangel TF, Bini LM (2013) Mantel test in population genetics. Genet Mol Biol 36:475–485
Dornhaus A, Powell S (2009) Foraging and defense strategies. In: Lach L, Parr CL, Abbott KL (eds) Ant ecology. Oxford University Press, Oxford, pp 201–232
Dray S, Dufour A-B (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20
Ellis S, Robinson EJH (2015) Internest food sharing within a wood ant colonies: resource redistribution behaviour in a complex system. Behav Ecol 27:660–668
Eriksson ES (1985) Attack behaviour and distance perception in the Australian bulldog ant Myrmecia nigriceps. J Exp Biol 119:115–131
Greiner B, Narendra A, Reid SF, Dacke M, Ribi WA, Zeil J (2007) Eye structure correlates with distinct foraging bout timing in primitive ants. Curr Biol 17:R879–R880
Guillot G, Rousset F (2013) Dismantling the Mantel test. Methods Ecol Evol 4:336–344
Hartig F (2020) DHARMa: residual diagnostics for hierarchical (multi-level/mixed) regression models. R package version 0.3.3.0. https://CRAN.R-project.org/package=DHARMa
Hasegawa E, Crozier RH (2006) Phylogenetic relationships among species groups of the ant genus Myrmecia. Mol Phylogenet Evol 38:575–582
Helms JA IV (2017) The flight ecology of ants (Hymenoptera: Formicidae). Myrmecol News 26:19–30
Hölldobler B, Wilson EO (1990) The ants. Harvard University Press, Cambridge, p 732
Jones O, Wang J (2010) COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 10:551–555
Kalendar R, Khassenov B, Ramankulov Y, Samuilova O, Ivanov KI (2017) FastPCR: an in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109:312–319
Kamhi JF, Barron AB, Narendra A (2020) Vertical lobes of the mushroom bodies are essential for view-based navigation in Australian Myrmecia ants. Curr Biol 30:3432–3437
Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee D, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buass FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJC, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MMW, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Manbati J (2011) DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Science 471:325–330
Kassambara A (2020) ggpubr: 'ggplot2' based publication ready plots. https://CRAN.R-project.org/package=ggpubr
Katzerke A, Neumann P, Pirk CWW, Bliss P, Moritz RFA (2006) Seasonal nestmate recognition in the ant Formica exsecta. Behav Ecol Sociobiol 61:143–150
Kellner K, Reeve HK (2014) Genetic variability, queen number, and polyandry in social Hymenoptera. Evolution 48:694–704
Kofler R, Schlötterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685
Krapf P, Russo L, Arthofer W, Möst M, Steiner FM, Schlick-Steiner BC (2018) An alpine ant’s behavioural polymorphism: monogyny with and without internest aggression in Tetramorium alpestre. Ethol Ecol Evol 30:220–234
Narendra A, Ribi WA (2017) Ocellar structure is driven by the mode of locomotion and activity time in Myrmecia ants. J Exp Biol 220:4383–4390
Narendra A, Reid SF, Hemmi JM (2010) The twilight zone: ambient light levels trigger activity in primitive ants. Proc R Soc B 277:1531–1538
Narendra A, Reid SF, Greiner B, Peters RA, Hemmi JM, Ribi WA, Zeil J (2011) Caste-specific visual adaptations to distinct daily activity schedules in Australian Myrmecia ants. Proc R Soc B 278:1141–1149
Narendra A, Greiner B, Ribi WA, Zeil J (2016) Light and dark adaptation mechanisms in the compound eyes of Myrmecia ants that occupy discrete temporal niches. J Exp Biol 219:2435–2442
Narendra A, Kamhi FJ, Ogawa Y (2017) Moving in dim light: behavioural and visual adaptations in nocturnal ants. Integr Comp Biol 57:1104–1116
Ogata K (1991) Ants of the genus Myrmecia Fabricius: a review of the species groups and their phylogenetic relationships (Hymenoptera: Formicidae: Myrmeciinae). Syst Entomol 16:353–381
Ogata K, Taylor RW (1991) Ants of the genus Myrmecia Fabricius: a preliminary review and key to the named species (Hymenoptera: Formicidae: Myrmeciinae). J Nat Hist 25:1623–1673
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Qian Z-Q, Ceccarelli FS, Carew ME, Schlüns H, Schlick-Steiner BC, Steiner FM (2011a) Characterization of polymorphic microsatellites in the giant bulldog ant, Myrmecia brevinoda and the jumper ant M Pilosula. J Insect Sci 11:71
Qian Z-Q, Schlick-Steiner BC, Steiner MF, Robson SKA, Schlüns H, Schlüns EA, Crozier RH (2011b) Colony genetic structure in the Australian jumper ant Myrmecia pilosula. Insectes Soc 59:109–117
Qian Z-Q, Schlüns H, Schlick-Steiner BC, Steiner FM, Robson SKA, Schlüns EA, Crozier RH (2011c) Intraspecific support for the polygyny-vs.-polyandry hypothesis in the bulldog ant Myrmecia brevinoda. Mol Ecol 20:3681–3691
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.r-project.org/
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Readhead R (2014) Chemical ecology and nest-mate recognition in the Australian bulldog ant Myrmecia nigriceps. (Unpublished honour's thesis). Deakin University, Australia
Reid SF, Narendra A, Taylor RW, Zeil J (2013) Foraging ecology of the night-active bull ant, Myrmecia pyriformis. Aust J Zool 61:170–177
RStudio Team (2020) RStudio: integrated development for R. RStudio, Inc. https://www.rstudio.com/
Sanetra M (2011) Nestmate relatedness in the Australian ant Myrmecia pyriformis SMITH, 1858 (Hymenoptera: Formicidae). Myrmecol News 15:77–84
Schlick-Steiner BC, Steiner FM, Sanetra M, Seifert B, Christan E, Stauffer C (2007) Lineage specific evolution of an alternative social strategy in Tetramorium ants (Hymenoptera: Formicidae). Biol J Lin Soc 91:247–255
Shattuck SO (1999) Australian ants: their biology and identification. CSIRO, Collingwood, p 256
Sheehan ZBV, Kamhi JF, Seid MA, Narendra A (2019) Differential investment in brain regions for a diurnal and nocturnal lifestyle in Australian Myrmecia ants. J Comp Neurol 527:1261–1277
Steiner FM, Crozier RH, Schlick-Steiner BC (2009) Colony structure. In: Lach L, Parr CL, Abbott KL (eds) Ant ecology. Oxford University Press, Oxford, pp 177–193
Taylor RW (2015) Ants with attitude: Australian jack-jumpers of the Myrmecia pilosula species complex, with descriptions of four new species (Hymenoptera: Formicidae: Myrmeciinae). Zootaxa 3911:493–520
Thurin N, Aron S (2008) Seasonal nestmate recognition in the polydomous ant Plagiolepis pygmaea. Anim Behav 75:1023–1030
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Microchecker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol 4:535–538
Van Wilgenburg E, Dand S, Forti AL, Koumoundouros TJ, Ly A, Elgar MA (2007) An absence of aggression between non-nestmates in the bull ant Myrmecia nigriceps. Naturwissenschaften 94:787–790
Ward PS, Brady SG (2003) Phylogeny and biogeography of the ant subfamily Myrmeciinae. Invertebr Syst 17:361–386
Wheeler WM (1916) The marriage-flight of a bull-dog ant (Myrmecia sanguinea F. Smith). J Anim Behav 6:70–73
Wickham H (2016) ggplot2: elegant graphics for data analysis. https://ggplot2.tidyverse.org
Acknowledgements
We thank the Australian Research Council for their Grants to AN (FT140100221, DP150101172); the University of Innsbruck/International relations office, for granting a KWA-scholarship to VA, and Suzanne Hodgkinson and Bruce Hall for their help and for accommodation; Zachary Sheehan and Duncan Jaroslow for their help with fieldwork, Philipp Andesner and Elisabeth Zangerl for their help in the lab, Théotime Colin for his advice and support, and all members of the Ecological Neuroscience Group at Macquarie University and the Molecular Ecology Group at the University of Innsbruck who helped with this work; Jason Brown for being a never-ending source of motivation; and two anonymous reviewers for constructive criticism of an earlier version of the paper.
Funding
Open access funding provided by University of Innsbruck and Medical University of Innsbruck. Grants by the Australian research council (FT140100221, DP150101172).
Author information
Authors and Affiliations
Contributions
AN, WA, FMS, and BCS-S designed the research; VA and AN did the fieldwork; VA and WA did the molecular-genetic analyses in the laboratory; VA, WA, PK, FMS, and BCS-S analyzed the data; VA wrote the manuscript, with contributions from AN, WA, PK, FMS, and BCS-S.
Corresponding author
Ethics declarations
Conflict of interests
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Als, V., Narendra, A., Arthofer, W. et al. Colony structure, population structure, and sharing of foraging trees in the ant Myrmecia nigriceps (Hymenoptera: Formicidae). Insect. Soc. 68, 327–335 (2021). https://doi.org/10.1007/s00040-021-00831-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00040-021-00831-7