Abstract
Dendritic cells (DCs) play a crucial role in orchestrating immune responses, particularly in promoting IFNγ-producing-CD8 cytotoxic T lymphocytes (CTLs) and IFNγ-producing-CD4 T helper 1 (Th1) cells, which are essential for defending against viral infections. Additionally, the nuclear envelope protein lamin A/C has been implicated in T cell immunity. Nevertheless, the intricate interplay between innate and adaptive immunity in response to viral infections, particularly the role of lamin A/C in DC functions within this context, remains poorly understood. In this study, we demonstrate that mice lacking lamin A/C in myeloid LysM promoter-expressing cells exhibit a reduced capacity to induce Th1 and CD8 CTL responses, leading to impaired clearance of acute primary Vaccinia virus (VACV) infection. Remarkably, in vitro-generated granulocyte macrophage colony-stimulating factor bone marrow-derived DCs (GM-CSF BMDCs) show high levels of lamin A/C. Lamin A/C absence on GM-CSF BMDCs does not affect the expression of costimulatory molecules on the cell membrane but it reduces the cellular ability to form immunological synapses with naïve CD4 T cells. Lamin A/C deletion induces alterations in NFκB nuclear localization, thereby influencing NF-κB-dependent transcription. Furthermore, lamin A/C ablation modifies the gene accessibility of BMDCs, predisposing these cells to mount a less effective antiviral response upon TLR stimulation. This study highlights the critical role of DCs in interacting with CD4 T cells during antiviral responses and proposes some mechanisms through which lamin A/C may modulate DC function via gene accessibility and transcriptional regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viruses exploit various entry routes to penetrate body barriers, infect diverse cell types, and adapt to evade immune surveillance. Eradicating rapidly replicating viruses effectively requires a multifaceted immune response orchestrated by different cellular entities targeting various viral components. Upon infection, the innate immune system promptly mobilizes to counteract viral dissemination, thus triggering the adaptive arm to initiate a specific immune defense, ultimately resulting in viral clearance [1].
The immunization with the Vaccinia virus (VACV), a linear double-stranded DNA virus, has shown efficacy against Variola major, contributing significantly to the global eradication of smallpox [2]. Notably, the priming, activation, proliferation, and differentiation of T cells in response to VACV rely on professional antigen-presenting cells (APCs), particularly dendritic cells (DCs) [3].
DCs can be categorized into distinct groups based on phenotype, function, and development [4, 5], including plasmacytoid DCs (pDCs), conventional DCs (cDCs) and, monocyte-derived DCs (moDCs). Under homeostasis, immature DCs are distributed throughout the body and at microbial entry points, serving as sentinels for danger signals. Common DC progenitors are the primary source of DCs. However, in mice, inflammation at infected sites triggers the recruitment of Ly6Chigh monocytes from the bloodstream [6]. Upon arrival, these monocytes differentiate into moDCs [7] in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) [8], which is produced by activated T cells and other cell types in the inflamed tissue [8]. In addition to tissue-resident DCs, moDCs initiate additional rounds of T cell priming [9].
Upon viral encounter in mice and humans, both peripheral tissue-resident DCs and moDCs recognize pathogens via germ-line encoded pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs) and cytoplasmic DNA sensors [10]. Among these, TLR signaling cascades, bifurcated into MyD88-dependent pathway and TRIF-dependent pathway. TLR4 signaling depends on both adaptor molecules, TRIF and MyD88, while TLR2 depends on MyD88 [11]. Upon detection of PAMPs, the Toll/Interleukin-1 receptor (TIR) domain-containing adaptor proteins, MyD88 and/or TRIF, are recruited to the TLRs. This initiation triggers signaling pathways that lead to the activation of NF-κB, interferon regulatory factors (IRFs), and mitogen-activated protein kinase (MAPK) cascades. These pathways ultimately induce the upregulation of costimulatory molecules (CD40, CD80, and CD86), inflammatory cytokines (such as IL-12, IL-6, and TNF), chemokines (RANTES, IP-10, ENA78, etc.), and type I interferons (IFNs) [12,13,14].
Maturation of both tissue-resident DCs and moDCs is imperative for effective priming of naïve CD8 and CD4 T cells, thereby initiating adaptive immunity [15] via major histocompatibility complex class I/II (MHCI/II) molecules to CD8 and CD4 T cells, respectively [16]. Upon antigen recognition, CD8 T cells differentiate into cytotoxic T lymphocytes (CD8 CTLs), which exert precise antiviral effects via cytokine secretion and release of cytotoxic granules [17, 18]. Similarly, CD4 T cells differentiate into T helper 1 cells (Th1), orchestrating antiviral responses through IFNγ production [18], engaging in direct cytotoxic mechanisms [18,19,20] and assist in B cell-mediated viral clearance via affinity maturation and antibody class switching [21]. Additionally, they contribute to CD8 T cell expansion and maintenance and memory against the virus [22, 23].
In mammalian cells, the nuclear envelope is composed of the nuclear pore complex, embedded in both the outer and inner nuclear membranes, along with the nuclear lamina. The nuclear lamina is composed of two main components, A- and B-type lamins, which contribute to the mechanical stability of the nucleus and regulate nuclear positioning, chromatin structure, the organization of the nuclear pore complex, the behavior of the nuclear envelope during mitosis, DNA replication, DNA damage responses, cell-cycle progression, cell differentiation, cell polarization during migration, and transcription [24, 25]. Our previous research has demonstrated the pivotal role of lamin A/C in enhancing T cell activation and promoting Th1 responses, which are crucial for effective viral clearance [20, 26, 27]. Furthermore, the absence of lamin A/C in T cells promotes their differentiation towards a Treg phenotype [28, 29]. Despite extensive information on the importance of lamin A/C in T cell immunity, its significance in DC functions, particularly in orchestrating the interplay between innate and adaptive immune responses to viral infections, remains an area that warrants further investigation.
This study provides valuable insights into the role of DCs in interacting with CD4 T cells during the antiviral response and the potential underlying molecular mechanisms controlled by lamin A/C, involving gene accessibility and transcriptional regulation in DCs.
Materials and methods
Mice
C57BL/6-Tg (TcraTcrb)425Cbn/J mice (CD45.1 OT-II mice) expressing a TCR specific for the OVA peptide (amino acid residues 323 to 339) in the context of I-Ab were obtained from the Jackson Laboratory (stock number 004194). The LysM-cre Lmnaflox/flox mouse (referred as LysM-Lmna−/−) was generated by crossing C57BL/6 LysM-cre+/− mice [30] with C57BL/6 Lmnaflox/flox mice [31]. C57BL/6 Lmnaflox/flox littermates were used as WT control mice. C57BL/6-CD45.2 and C57BL/6-CD45.1 WT mice were used as recipients for adoptive transfer. Lmnaflox/flox mice were generously provided by Y. Zheng [31]. All mice were bred at the CNIC under specific pathogen-free conditions. Experiments were performed using mice that were matched for both sex and age (6–12 weeks). The mice were maintained on a standard 12-hour light/dark cycle (with the light period from 7 a.m. to 7 p.m.), and they had access to food and water ad libitum. The Animal Care and Ethics Committee of the CNIC, UAM, and the regional authorities approved all experimental procedures.
Antibodies and reagents
Antibodies against: CD4 (Clone RM4-5, GK1.5) -v450, -APC and PE; CD25 (PC61.5) -APC; CD69 (H1.2F3)-FITC; CD28 (37.51); CD3 (145-2C11); CD45.1 (A20)-Pe-Cy7, -v450, -FITC; CD45.2 (104) -v450, -redFluor710; CD11b (M1/70)-v450; CD86 (GL-1) PE, PE-Cy7; IFNγ XMG1.2 FITC, -APC; Foxp3 (3G3)-FITC; MHC-II (I-A/I-E, M5/115.15.2)-APC; GR1 (RB6-8C5)-Biotin; CD80 (16-10A1)-Biotin, -VB711; B220 (RA3-6B2)-Biotin; F4/80 (BM8.1)-Biotin; CD11b (M1/70)-Biotin, -V450; CD19 (1D3)-Biotin; CD25 (PC61.5)-Biotin from Tonbo. CD80 (16-10A1)-VB711; CD11c (N418)-FITC MHC-II (M5/114.15.2)-AlexaFluor 700; IL-6 (MP5-20F3)-APC; TNFα (MP6-XT22)-PerCP-Cy5.5; CD40 (3/23) PE-CF594 from BioLegend; CD86 (GL1) -PE-Cy7 from eBioscience. Lamin A/C (4C11)-Alexa488; NF-κB (D14E12-XP) from Cell Signaling. IgM (R6-60.2)-Biotin; MHC-II (2G9)-Biotin; CD49b (DX5)-Biotin; Streptavidin-PE from BD Bioscience. Lipopolysaccharide (LPS) (from Sigma-Aldrich, Catalog number L2630, final concentration 20 ng/ml); Palmitoyl-3-CysSerLys-4 (Pam3CSK4) (InvivoGen, tlrl-pms, 100 ng/ml); 7-amino-4 chloromethylcoumarin (CMAC) CellTrakerTM (Invitrogen, C2110, 0.1 µM); Vybrant CFDA SE Cell Tracer Kit-Carboxyfluorescein diacetate succinimidyl ester (CFSE) (Invitrogen, V12883, 5 µM); CellTrace Violet (CTV) (Invitrogen, C34557 5 µM); Cell Stimulation Cocktail (500X) (Tonbo, TNB-4975-UL100, 1X); 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (Invitrogen, D1306, 1:10000); Vybrant DiL Cell-Labeling Solution (DiL tracer) (Invitrogen, V22885, 1:1000); Mouse GM-CSF (PeproTech, 315-02, 20 ng/ml); OVA-OT-IIp(OVA323-339, ISQAVHAAHAEINEAGR) (GenScript, 10 µg/ml); Fluorescence Mounting Medium (Dako Omnis) with 500 µg/l DAPI (Agilent, GM30411-2, 30 µl/sample); SYBR Green PowerUp master mix (Cat. #A25742, Applied Biosystems); Spleen dissociation Medium (Cat. #07915, Stem Cell), EasySep Mouse CD11c Positive Selection Kit II (Cat #18780, Stem Cell).
T cell isolation
Spleen and LN cell suspensions from CD45.1 OT-II mice were obtained by homogenization through a 70 μm cell strainer (Falcon). In spleen suspensions, red blood cells (RBC) were lysed using eBioscience Red Blood Cell Lysis Buffer (Invitrogen) for 5 min at RT. Naïve CD4 T cells were isolated using the EasySep Kit or negative selection with labeled antibodies (anti GR-1, B220, F4/80, CD8α, CD11b, CD19, CD25, IgM, MHC-II and CD49b) and streptavidin-bound magnetic microbeads (Miltenyi Biotec). Isolated CD4 T cells were activated with anti-CD3 and anti-CD28 antibodies if needed.
Generation of bone marrow-derived DCs
Bone marrow-derived DCs (BMDCs) were generated as described by [32]. Briefly, femur and tibiae were extracted from 6 to 12 weeks old WT and LysM-Lmna−/− mice. After removing surrounding muscle tissue, both bone ends were cut, and marrow was flushed with PBS using a 25G syringe. RBCs were lysed with eBioscience Red Blood Cell Lysis Buffer. The bone marrow cell suspension was cultured on non-treated 150-mm Petri dishes at a concentration of 0.5 × 106 cells/ml in complete RPMI-1640 with 20 ng/ml rGM-CSF (PeproTech). On day 3, attached cells were removed, and the remaining cells were cultured until days 7–9. Maturation was induced based on the experiment, either with an overnight incubation with TLR4 activator Lipopolysaccharide (LPS), 4-hour treatment with TLR2 activator Pam3CsK4, or with irradiated VACV extract (stock 107 p.f.u, use 1:1000) at 37 ºC. On days 7–9, BMDCs were collected and used for subsequent experiments.
BMDCs and CD4 T cells in vitro co-cultures
Naïve CD4 T cells were isolated from CD45.1 OT-II mice, and GM-CSF-BMDCs were matured with LPS (overnight), Pam3CsK4 (4 h), or irradiated VACV extract (4 h), followed by pulsing with 10 µg/ml OVA323–339 peptide (OVA-OT-IIp) for at least 30 min at 37 ºC. Simultaneously, naïve CD4 T cells were labeled with CellTrace Violet following the manufacturer’s protocol (Life Technologies). Co-cultures of BMDCs and CD4 T cells were conducted with a 1:2 ratio, respectively, in 96-well U-bottom plates. CTV-labeled CD4 OT-II T cells cultured alone served as a negative control, and CD4 OT-II T cells stimulated with plate-bound anti-CD3 (5 µg/ml; Tonbo) and soluble anti-CD28 (1 µg/ml; Tonbo) antibodies served as positive controls. Activation, proliferation, and differentiation of naïve CD4 OT-II T cells in response to OVA323–339 peptide-primed BMDCs subsets were assessed by flow cytometry, monitoring specific markers (CD25 and CD69 for activation, IFNγ for Th1 differentiation, and IFNγ-producing CD8 T cells) and the reduction in CTV intensity at 24 h, 48 h, and 6 days.
BMDCs and CD4 T cells in vivo co-cultures
For adoptive transfer experiments, CD45.1 OT-II recipient mice received a subcutaneous injection (s.c.) in footpad with 2 × 106 WT- or Lmna−/− GM-CSF-BMDCs matured with LPS and pulsed with OVA323–339 cognate OT-IIp. Six days after BMDC injection, draining popliteal lymph nodes (pLN) were extracted, and Th1 differentiation was assessed by flow cytometry.
Flow cytometry
T-cells were stimulated for 6 h with PMA (20 ng/ml) plus ionomycin (1 µg/ml). Brefeldin A (5 µg/ml Sigma-Aldrich) was added for the last 2 h to allow intracellular cytokine accumulation. Single-cell suspensions were transferred to a 96-V-well plate and pre-incubated with a purified anti-CD16/32 antibody (FcBlock, Tonbo) for 10 min on ice before immunostaining. Cells were then stained with the appropriate surface marker antibodies, on ice for at least 30 min according to the manufacturer’s instructions. Finally, a viability marker was added to the labeled cell suspension. To assess IFNγ production intracellularly, CD4 T cells were stimulated with Cell Stimulation Cocktail (500X) (Tonbo) for 4 h at 37 ºC to allow intracellular cytokine accumulation. After surface staining, samples were fixed and permeabilized with a Cytofix/Cytoperm kit (BD Biosciences) for 20 min at 4 ⁰C. The solution was washed out with Perm/Wash™ Buffer (BD Biosciences), and then, cells were intracellularly stained with anti–IFNγ for 1 h at RT. For Foxp3 staining, the mouse Foxp3 Buffer set (BD Pharmigen) was used following the manufacturer’s protocol. For lamin A/C intranuclear staining, cells were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.5% Triton X-100/PBS solution for 5 min at RT. Anti-mouse lamin A/C antibody was then incubated for 1 h at RT. Data were acquired on LSRFortessa or FACSCanto flow cytometers (BD Biosciences) and analyzed with FlowJo V10.4.2 software (Treestar Inc.).
Time-lapse fluorescence microscopy
For analysis of T cell-BMDC cognate interactions, WT or Lmna−/−-BMDCs matured with LPS for 24 h and pulsed with OVA323–339 cognate OVA-OT-IIp were mixed with GFP-CD4 OT-II T cells (1:1). To observe live T cell-BMDC interactions, BMDCs labeled with DiL tracer (Vybrant™ DiI Cell-Labeling Solution, Invitrogen) were suspended in Hanks balanced salt solution (HBSS) (Lonza) with 2% FBS and 20 mM HEPES (Hyclone) and plated onto poly-L-Lysine (PLL; 20 µg/ml, Sigma-Aldrich)-coated 35-mm-diameter culture dishes (MatTek) for at least 15 min at 37 °C. After BMDCs adhered to the PLL coating, CD4 T cells were added to the BMDC culture and placed into microscope (Nikon ECLIPSE Ti-TimeLapse microscopy), covered by an acrylic box for acquisition at 37 °C and 5% CO2. T cell-BMDC interactions were recorded for 150 min. Image analysis was performed using Imaris Software (Bitplane). The software generates individual volumes of T cells and BMDCs based on intensity and size. It automatically measures the number of cells, their localization, and the speed of T cells during the recording. To determine the distance from a T cell to the nearest BMDC, the software creates a new channel that represents a gradient of intensities proportional to the distance to the nearest BMDC. The mean intensity within the T cell’s volume in this new channel indicates the distance to the nearest BMDC.
Fluorescence confocal microscopy
For immunostaining, WT- or Lmna−/−-BMDCs were treated or not with LPS for 24 h and then plated onto PLL-coated coverslips (20 µg/ml) for 30 min at 37 °C. Subsequently, cells were fixed in 4% PFA, permeabilized in 0.5% Triton X-100/PBS solution for 5 min, and stained with appropriate antibodies: anti-NF-κB p65 and Alexa488-conjugated anti-mouse lamin A/C antibodies (both from Cell Signaling) and DAPI (Invitrogen) for 1 h at RT. Finally, samples were mounted in Prolong antifade medium (Invitrogen). Confocal images were acquired using a Leica gated STED-3X-WLL SP8 microscope under the same conditions, including identical exposure times and light intensity. Image analysis was performed using ImageJ Fiji software and Imaris software (Bitplane).
Reverse transcription-quantitative PCR
Total RNA from cultured WT or Lmna−/−-BMDCs (treated with LPS for 24 h) was isolated with TRI Reagent solution (Invitrogen) and isopropanol precipitation. RNA purity and concentration were assessed from the ratio of absorbance at 260 and 280 nm in a Nanodrop-1000 Spectrophotometer (Thermo Scientific). Total RNA (0.5 to 2 µg) was reverse transcribed to complementary DNA (cDNA) with the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems). Quantitative PCR was performed using the PCR Power SYBR Green PCR Master Mix (Applied Biosystems) in a B7900-FAST-384 Sequence Detection System (Applied Biosystems), with technical triplicates. Expression levels of target genes were normalized to some of the housekeeping genes: β-actin, Ywhaz (Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein Zeta), or Rna18s1. Gene-specific primers used, and their sequences are listed in Table 1.
Vaccinia virus (VACV) in vivo infection and viral titration
The VACV strain Western Reserve (WR, ATCC number VR-1354) was kindly provided by Jonathan W. Yewdell and Jack R. Bennink (NIH, Bethesda, Maryland, USA). VACV was propagated in African Green Monkey Kidney Fibroblast Cells (CV-1) and used as clarified sonicated cell extracts. WT or OVA-expressing VACV were administered by intraperitoneal (i.p.) injection, subcutaneous (s.c.) inoculation in the footpad [33] or by skin scarification (s.s.) in the tail of WT and LysM-Lmna−/− mice. 1 × 105 or 1 × 106 plaque-forming units (p.f.u.) were used depending on the route of administration. For this last procedure, animals were anesthetized with inhalatory isoflurane, and then, thirty scarifications were made with a 25 G syringe in 1 cm along the base of the tail avoiding bleeding, as previously described [34]. After that, 1 ml of PBS containing 1 × 106 p.f.u. of VACV-WR was added to the area and allowed to air dry. Only PBS was added to the mock-infected animals. For viral quantification, mice were euthanized, and the tail was aseptically removed, weighed, frozen, and stored at -80 ºC until usage. The samples were homogenized with a Tissue Ruptor (QIAGEN, USA) in 0.5 ml DMEM containing 50 U/ml penicillin and 100 µg/ml streptomycin. The homogenates were sonicated for 3 min at 40% amplitude, freeze-thawed twice (-80 ºC/ 37 ºC), sonicated again under the same conditions, and then serially diluted in DMEM without FBS. To quantify p.f.u., dilutions were added to monolayers of CV-1 seeded on 24-well plates and incubated at 37 ºC and 5% CO2 for 1 h. Hereafter, 0.5 ml of DMEM containing 0.5% FBS was added to each well. After 24 h, cells were stained for 5 min with crystal violet solution (0.5% crystal violet, 10% ethanol, and 1% PFA) and washed again. Viral plaques were counted, and data was presented as p.f.u./g [35].
Isolation of CD11c+ cells
Spleens were digested with spleen dissociation medium following manufacturer instructions (#07915, Stem Cell) for 30 min at RT. Red blood cells were lysed and the resulting splenic cell suspensions were resuspended in PBS containing 5 mM EDTA, 0.5% BSA, and 5 µg/ml DNase-I. CD11c+ cells were isolated using the EasySep Mouse CD11c Positive Selection Kit II (Stem Cell). The CD11c+ splenic cells were then incubated in vitro with 20 ng/ml LPS for 5 h with the addition of 5 µg/ml Brefeldin A during the final 4 h. Cell subsets were analyzed using flow cytometry by staining with fluorescence antibodies against CD11c, CD11b, CD80, CD86, MhcII, CD40 prior to fixation, and against IL-6 and TNFα after fixation and permeabilization (all Abs from BD Biosciences, Cell Signaling and ABCAM).
ATAC-seq
50.000 CD11c+ BMDCs were treated with LPS, isolated, and cryopreserved in 10% DMSO + 90% SF (n = 3 for each condition). After rapid thawing, cells were counted, lysed (10mM PIPES pH 6.8, 100mM NaCl, 300mMsucrose, 3mMMgCl2, 0.1% TritonX-100) for 5 min at 4 ºC and centrifuged at 800 g. Supernatant was discarded, and nuclei were resuspended in transposase reaction buffer with transposase5 (1 uL Tn5/10.000 cells) and incubated for 30 min at 37 ºC. Later, SDS at a final concentration of 0.1% was added to stop the reaction (30 min, 40 ºC) and DNA was isolated with KAPA Pure beads (SPRI beads). Briefly, lysates were incubated with SPRI beads (2x), placed on the magnet, washed twice with 70% ethanol, and air-dried. DNA was eluted with elution buffer (EB) and its concentration measured with Qubit kit (Invitrogen, Waltham, MA, USA). For DNA library generation, qPCR with i5 and i7 primers (NEXTERA barcoding) was performed (98 ºC 2 min, (98 ºC 20 s, 63 ºC 30 s, 72 ºC 1 min); 14 cycles). Then, a size cut-off was conducted. First, amplified DNA was incubated with 0.5x concentration of SPRI beads. Then, beads were discarded, and supernatant was incubated with 2x concentration of SPRI beads. Beads were washed twice with 70% ethanol, eluted with EB, and measured with Qubit. Finally, Agilent HS Bioanalyzer was used to assess peak sizes and percentages.
Libraries were sequenced on a HiSeq 4000 to generate paired reads and processed with RTA v1.18.66.3. FastQ files for each sample were obtained using bcl2fastq v2.20.0.422 software (Illumina, San Diego, CA, USA).
Bioinformatics analysis
Read quality was assessed using FastQC. Sequencing reads were trimmed for Illumina adapter and transposase sequences with cutadapt and were aligned to the mouse reference genome mm10 (GRCm38) with bowtie aligner with a limit of 1000 bp paired distance. Then, aligned reads were cleaned of duplicates reads with samtools MarkDuplicates tool. Peaks were identified with HOMER2 in region mode, with a local size of 50,000 bp, a peak expected size of 150 bp, a minimum non-unified distance of 1000 bp, a minimum coverage of 5 reads and a region resolution of 6. For each condition, peaks detected in all replicates were taken into account. For the final peak count, all peaks detected in any of the WT/Lmna−/− conditions were used to define a consensus of 21,107 well defined peaks. Peaks were also annotated with HOMER2. Peak coverage and differences between WT and Lmna−/− were calculated with HOMER2 and DESeq2. Bigwig files for visualization were created with Deeptools and Wiggletools. Tracks were represented in IGV browser. Final aggregated figures were represented with custom Python scripts. Over-representation analysis and their figure representation were done with clusterProfiler v4.4.2 [36], ReactomePA v1.34.0 [37] and enrichplot v1.10.2 R packages. Data available in GEO under accession number GSE269613.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 8 software. Normality of the data was tested using the Kolmogorov-Smirnov test. The parameters followed a normal distribution, and, therefore they were represented as the distribution of each sample, mean ± SEM (Standard Error of the Means). Parametric tests were used for statistical analysis. Unless otherwise stated, statistical significance was calculated by Unpaired Student’s t-test. When specified, one-way ANOVA or two-way ANOVA were used, with Bonferroni’s post-hoc multiple comparison test applied as appropriate. The significance of differences was calculated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001. Not significant was depicted as n.s.
Results
Reduced lamin A/C levels in the myeloid compartment partially impede the response to VACV by affecting T cell-mediated stimulation
The importance of lamin A/C within the myeloid compartment and the functional role of in vitro generated GM-CSF BMDCs were evaluated through various in vivo and in vitro infection and inflammation approaches.
To selectively downregulate lamin A/C expression in specific myeloid cells, including moDCs and in vitro generated-BMDCs, we established LysM-cre+/− Lmnaflox/flox mice referred to as LysM-Lmna−/− or Lmna−/− hereafter. LysM-Lmna−/− mice exhibited physical characteristics and body weight comparable to wild-type (WT) mice with no discernible differences in the size, weight, and morphology of various immune system organs, such as the spleen or thymus, when compared to WT mice (described in [38]). Immunofluorescence analysis of cell populations in homeostasis in organs such as the spleen, mesenteric lymph nodes (mLN), lung, or bone marrow (BM) revealed no notable differences between genotypes, except for a reduction in the percentage of alveolar macrophages in adult LysM-Lmna−/− animals within the studied populations [38].
To explore the impact of lamin A/C in the myeloid compartment in the response to the virus, mice were infected with VACV, known for inducting robust Th1 immune response in humans [39] and in mice [1, 20, 40]. Given the role of CD4 T cells in regulating the primary CD8 T cell response to VACV infection depending on the route of infection and viral dose [41, 42], various administration routes were employed including subcutaneous (s.c.), inoculation in the footpad, intraperitoneal injection (i.p.), or via skin scarification (s.s.) in the tail vein (Fig. 1).
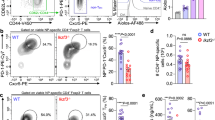
Lamin A/C modulation in myeloid compartment influences vaccinia virus susceptibility and protection across infection routes. (A) Subcutaneous inoculation (s.c.) with 105 p.f.u. of VACV was administered to both WT and LysM-Lmna−/− mice. Flow cytometry analysis of IFNγ-producing CD4 and CD8 T cells, as well as CD25+Foxp3+ Treg cells, was conducted in popliteal lymph nodes (pLN) at 6- and 14-days post-infection. The percentage of Th1 cells, CD8 CTL T cells, and Treg cells was assessed. The data represent means ± SEM from at least 8 mice per group, obtained from 3 independently conducted experiments, and were analyzed using unpaired Student’s t-test between genotypes at each time point. Statistical significance is denoted by asterisks (*P < 0.05; **P < 0.01; ns, not significant). (B) Intraperitoneal inoculation (i.p.) with 106 p.f.u. of VACV was performed on both WT and LysM-Lmna−/− mice. Flow cytometry analysis of IFNγ-producing CD4 and CD8 T cells was conducted in mesenteric lymph nodes (mLN) 6 days post-infection. The percentage of Th1 and CD8 CTL cells was determined. Data represent means ± SEM of at least 6 mice per group, derived from 2 independently conducted experiments, and were analyzed by unpaired Student’s t-test. Statistical significance is indicated by asterisks (*P < 0.05; **P < 0.01). (C) Infection with 105 p.f.u. of VACV through skin scarification (s.s.) in the tail was performed on WT and LysM-Lmna−/− mice. Flow cytometry analysis of the percentages of IFNγ-producing CD4 and CD8 T cells was conducted in extracted inguinal lymph nodes (iLN) at 6- and 14-days post-infection. (D) Viral load measurement in the tail skin was performed at 6- and 14-days post-infection. The data represent means ± SEM of at least 4 mice per group, derived from 2 independently conducted experiments, and were analyzed by unpaired Student’s t-test. Statistical significance is denoted by asterisks (*P < 0.05; **P < 0.01; ns, not significant). This schematic was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution CC BY 4.0 License; https://smart.servier.com
In the initial infection experiments, both WT and LysM-Lmna-/- mice were administered VACV via the footpad (s.c.). Subsequently, at 6 and 14 days post-infection, popliteal lymph nodes (pLN) were harvested for the analysis of T cell populations (Fig. 1A). The frequencies of Treg cells and IFNγ-producing CD4 and CD8 T cells were assessed using flow cytometry. Only at the 6-day post-infection time point were differences observed between genotypes in the percentage of IFNγ-producing CD4 and CD8 T cells. No disparities were noted at the 14-day time point or in the percentage of Treg cells at either of the two time points analyzed.
Further experiments were conducted by inoculating WT and Lmna−/− mice i.p. with VACV (Fig. 1B). Six days later, the frequency of IFNγ-producing CD4 and CD8 T cells was analyzed in mLN using flow cytometry. Interestingly, in the mLN of LysM-Lmna−/− mice, there was a reduced percentage of both IFNγ+ CD4 and CD8 T cells at 6 days.
Similar outcomes were observed in mice administered VACV through s.s. in the tail skin (Fig. 1C). In this context, the percentage of IFNγ+ CD4 and CD8 T cells in the inguinal lymph nodes (iLN) was evaluated at two distinct time points: 6 and 14 days. While no statistically significant differences were noted at 6 days in IFNγ+ CD8 T cells, there was a significant reduction in IFNγ-producing CD4 T cells (Fig. 1C). By 14 days post-infection, a reduced number of both IFNγ-producing CD4 and CD8 T cells was observed in Lmna−/− mice (Fig. 1C). This trend correlated with viral titers at the infection site. We observed that VACV levels in the skin of infected mice were higher at 6 days than at 14 days post-infection through a s.s. route in WT mice. Interestingly, the virus levels at 6 days did not differ between WT and Lmna−/− mice. However, in WT mice, there was a subsequent reduction in virus levels in the skin by 14 days. This reduction was significantly less pronounced in Lmna−/− mice, resulting in higher virus levels in the skin at 14 days in Lmna−/− mice compared to WT mice (Fig. 1D). These findings underscore the importance of lamin A/C in myeloid cells for efficient virus clearance, particularly by influencing the stimulation of the adaptive immune system, specifically the Th1-cell-mediated and CD8 cytotoxic T lymphocyte (CTL) responses.
Deficiency of lamin A/C in GM-CSF BMDCs influences impaired T cell responses across maturation and stimulation environments
To resemble conditions similar to in vivo mo-DCs, bone marrow cells were cultured with GM-CSF [43]. The influence of lamin A/C on GM-CSF BMDCs (referred as BMDCs hereafter) in T cell activation, proliferation, and differentiation was then assessed under varied conditions. In a setup like VACV infection, BMDCs were exposed to irradiated VACV and then loaded with ovalbumin (OVA) peptide OVA323–339 (OVA-OT-IIp). These cells were co-cultured with naïve CD4 OT-II T cells, which express a TCR recognizing the OVA-OT-IIp, for 24–48 h for activation analysis and for 6 days for proliferation and differentiation towards the Th1 phenotype analysis, respectively. Lmna−/−-BMDCs co-cultures consistently showed a lower percentage of activated CD4 T cells at all observed time points, shown by decreased membrane expression of CD69 and CD25 in CD4 T cells (Fig. 2A). Additionally, these CD4 T cells displayed reduced proliferation capacity and diminished differentiation toward the Th1 phenotype, reflected in a reduction in the percentage of cells expressing IFNγ (Fig. 2A).
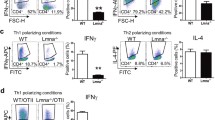
Lamin A/C deficiency in matured GM-CSF-BMDCs reduces CD4 T cell activation, proliferation and Th1 differentiation in vitro and in vivo. Flow cytometry was employed to assess the percentage of activated CD4 T cells, specifically gated on CD25+ or CD69+ cells; Th1-differentiated cells, specifically gated on IFNγ + cells; and proliferating CellTrace Violet+ T cells. Maturation of GM-CSF-derived WT- and LysM-Lmna−/−-BMDCs was undertaken with (A) sonicated VACV extract, (B) Pam3CSK4, or (C) lipopolysaccharide (LPS), and, in all conditions, pulsed with OVA cognate OT-IIp. (D) WT- or Lmna−/−-BMDCs, matured with LPS and pulsed with the OVA323–339 cognate OT-II peptide, were s.c. inoculated into CD45.1 OT-II recipient mice. Six days later, pLNs were extracted, and IFNγ-producing CD4 T cells were analyzed by flow cytometry. The data represent means ± SEM and are presented from a representative experiment out of 3 (A and B, n = 3) or out of 4 (C, n = 4) or out of 2 (D, n = 4). Activation data were analyzed using One Way ANOVA and Bonferroni post-test, and proliferation and Th1 differentiation were assessed by unpaired Student’s t-test. Statistical significance was denoted by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). This schematic was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution CC BY 4.0 License; https://smart.servier.com
In the immune system, the detection of viruses by DCs/APCs is not solely dependent on nucleic acid–specific receptors. Instead, some TLRs, such as TLR2 and TLR4, which do not recognize nucleic acids, play a crucial role in recognizing various viruses. This recognition is essential for distinguishing between different classes of pathogens to facilitate an appropriate immune response [44].
Therefore, given that VACV influences DCs, in part through TLR2 [45, 46] and TLR4 [47, 48], we stimulated BMDCs with TLR2 ligand Pam3CSK4 and TLR4 ligand LPS. Subsequently, BMDCs were pulsed with OVA-OT-IIp and co-cultured with naïve CD4 OT-II T cells for 24–48 h, and for 6 days. Upon TLR2 stimulation (Fig. 2B), Lmna−/−-BMDCs consistently induced a lower percentage of activated CD4 T cells at 48 h. Furthermore, these CD4 T cells displayed a diminished capacity to differentiate toward the Th1 phenotype, with no observed differences in proliferation rates between genotypes. In the context of LPS-induced maturation for TLR4 stimulation, Lmna−/−-BMDCs exhibited a lower percentage of activated CD4 T cells at 24 and 48 h. Importantly, these CD4 T cells also showed reduced proliferation and differentiation toward the Th1 phenotype (Fig. 2C).
To assess the stimulatory potential of these BMDCs within a more physiological context, in vivo adoptive transfer experiments were conducted. CD45.1 OT-II mice received s.c. injection in footpad with BMDCs matured with LPS and loaded with OVA-OT-IIp. Six days post-BMDCs injection, draining pLN were harvested, and Th1 differentiation was evaluated. Interestingly, recipient mice inoculated with Lmna−/−-BMDCs exhibited lower Th1 differentiation, as indicated by a reduced percentage of IFNγ+ CD4 T cells (Fig. 2D). These results indicate that the modulation of lamin A/C in BMDCs significantly impacts T cell responses, leading to decreased activation, proliferation, and differentiation, particularly under various maturation and stimulation conditions. This influence is probably mediated through mechanisms associated with TLR2 or TLR4 signaling pathways.
Lamin A/C in BMDCs controls the formation of immune synapse
Since some studies suggest that TLR2 might recognize virus preparations in vitro but plays a minor role in preventing VACV dissemination following systemic infection with large viral doses [49], and considering that TLR2 activation by VACV might be linked to Th2 conversion and robust humoral responses [50], which contrasts with the Th1 and CTL response observed in our in vivo studies associated with LPS stimulation [20], we explore the impact of lamin A/C on TLR4 signaling by stimulating with LPS. During antigen recognition, T cells and DCs establish communication through the immune synapse (IS), which is characterized by the precise redistribution and interaction of the TCR/CD3 at the T cell side and the MHC-II at the APC side, along with costimulatory and adhesion molecules on both sides [51]. Lamin A/C is crucial for TCR-dependent signaling and cytoskeletal reorganization at the T cell side [26, 27, 29]. To investigate this on the APC side, time-lapse fluorescence microscopy experiments were conducted. CD4+ T cells move within the lymph nodes, interacting with APCs until they recognize an antigen presented in the context of MHC-II. Upon antigen recognition, they form either short- or long-term contacts, which reduce their speed and proximity to the APCs. In vitro, the interaction between T cells and APCs can be quantified by measuring the speed of the T cells and their distance to the nearest APC (Supplementary Fig. 1). Analysis of the interaction between OT-II CD4+ T cells and LPS-matured OVA OT-IIp-loaded WT- or Lmna−/−- BMDCs revealed that CD4 T cells exhibited higher speed and increased distance to the nearest BMDC when co-cultured with Lmna−/−-BMDCs (Fig. 3A). This suggests a reduced ability of LPS-matured Lmna−/−- BMDCs to form stable conjugates, which may contribute to reduced activation, proliferation, and differentiation of T cells toward a Th1 phenotype [26, 27].
Reduced lamin A/C levels in matured GM-CSF-BMDCs diminish the formation of conjugates with T cells, without affecting the gene expression or membrane localization of MHC-II and costimulatory molecules. (A) WT- and Lmna−/−-BMDCs, matured with LPS and pulsed with OVA323–339 cognate OT-IIp, were co-cultured with GFP-OT-II CD4 T cells in a 1:1 ratio. Prior to the co-culture, BMDCs were stained with DiL and plated onto poly-L-Lysine. The study evaluated the speed of T cells (µm/s) and distance between T and BMDC (A.U.) at various time points. The presented data are from a representative experiment out of two. Each symbol in the upper graphs represents the mean speed and SEM of individual T cells over two different intervals (n ≥ 250), while in the lower graph, it represents the mean distance and SEM to the nearest BMDC of the detected T cells at the given time point (n > 250). The speed analysis was performed using an unpaired Student’s t-test while distance analysis was conducted using a two-way ANOVA and linear regression. Statistical significance is indicated by ***P < 0.001. A.U. denotes arbitrary units. (B) RT-qPCR analysis was performed on indicated genes from WT- and Lmna−/−-BMDCs, both stimulated with LPS for 24 h. The data, obtained from three independent experiments with 3 samples each, revealed no significant differences in the expression of Mhc-ii and costimulatory molecules genes between the two genotypes (n = 3). The analysis employed unpaired Student’s t-test, and the housekeeping genes Ywhaz, β-actin, and Rna18s1 were used for normalization. The data represent means ± SEM. (ns, not significant). (C) The membrane levels of MHC-II, CD80, and CD86 were evaluated on CD11c+ cells derived from WT and Lmna−/−-BMDCs, either matured or not with LPS. The geometric mean fluorescence intensity (MFI) of MHC-II+, CD80+, and CD86+ markers was analyzed. The data, representing means ± SEM from three independently conducted experiments (n = 3), underwent statistical analysis using one-way ANOVA with Bonferroni’s multiple comparison test. Significance levels are denoted by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). This schematic was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution CC BY 4.0 License; https://smart.servier.com
Given the observed disparities in the ability of BMDCs to form T cell conjugates, we investigated whether BMDCs exhibited distinct expression levels of MHC-II and costimulatory molecules, including Cd80, and Cd86. mRNA analysis showed no discernible differences between genotypes in LPS-matured BMDCs (Fig. 3B). The maturation process of BMDCs involves an upregulation of membrane expression levels of MHC-II, CD80, and CD86, which enhances T cell stimulation [52]. To assess their membrane expression by flow cytometry, we examined both LPS-untreated and LPS-treated matured BMDCs (Fig. 3C). As anticipated, there was a noticeable increase in the levels of costimulatory molecules associated with maturation. It is noteworthy that no significant differences were observed in the expression levels between genotypes in unmatured and LPS-matured conditions.
Lamin A/C in certain splenic CD11c + cell populations regulates MHC-II and co-stimulatory molecule expression, IL-6, IL-1α and TNFα levels
To investigate whether the effects of lamin A/C observed in LPS-matured BMDCs also occur in specific subpopulation of DCs in the mouse spleen, splenic CD11c+ cells were isolated from WT and LysM-Lmna−/− mice using antibody-coated microbeads for positive selection. Following isolation, these cells were stimulated with LPS. After stimulation, mRNA was extracted from a portion of the cells and quantified using RT-qPCR to analyze the levels of Il12bIl1a and Tnfa mRNA. Another portion of the isolated cells underwent immunofluorescence and flow cytometry to measure the membrane expression of MHC-II and co-stimulatory molecules, as well as the intracellular levels of IL-6 and TNFα. Under untreated conditions, no differences in the mRNA levels of Il12b,Il1a, and Tnfa were observed. However, LPS treatment induced the expression of Il12b, Il1a, and Tnfa mRNA in both WT and LysM-Lmna−/− cells. Notably, there was a significant reduction in the expression of Tnfa and Il1a mRNA in LysM-Lmna−/− cells compared to WT cells, while Il12b mRNA expression showed no significant difference (Fig. 4A). Isolated CD11c+ cells exhibited distinct levels of CD11b+, indicating the presence of various CD11c+ cell subpopulations in the spleen. We examined the membrane levels of MHC-II and co-stimulatory molecules, and the intracellular levels of TNFα and IL-6, in three different populations: CD11c++CD11b−, CD11c++CD11b+ and CD11c+CD11b++ cells, following ex vivo LPS stimulation. In CD11c++CD11b− cells, no differences were observed between WT and LysM-Lmna−/− cells regarding the percentage of positive cells or the fluorescence intensity of MHC-II, CD40, CD80, CD86, IL-6, and TNFα. CD11c++CD11b+ LysM-Lmna−/− cells exhibited increased TNFα+ cells compared to WT cells. Notably, CD11c+CD11b++ LysM-Lmna−/− cells showed reduced percentages of MHC-II+ and IL-6+ cells, as well as decreased fluorescence intensity for MHC-II, CD40, CD86, IL-6, and TNFα in positive cells (Fig. 4B). These findings indicate that lamin A/C modulation in specific splenic CD11c+ cell populations significantly affects the expression of MHC-II and certain co-stimulatory molecules, and pro-inflammatory cytokines.
Percentage of positive cells, membrane expression of MHC-II and co-stimulatory molecules, and intracellular levels of TNFα and IL-6 in specific populations of splenic CD11c+ cells in LysM-Cre Lmna−/− mice. (A) RT-qPCR analysis was performed on the indicated genes from isolated splenic CD11c+ WT- and Lmna−/− cells stimulated with LPS for 5 h. The data revealed significant differences in the expression of Tnfa and Il1a but not Il12b upon LPS stimulation between the two genotypes (n = 3–4 samples from two independent experiments). One-Way ANOVA and Tukey post-test were used for analysis, with Hprt1 and Rna18s1 as housekeeping genes for normalization. Data represent means ± SEM. (*P < 0.05; **P < 0.01). (B) Immunofluorescence staining and flow cytometry analysis was performed on LPS-treated splenic CD11c+ cells from WT and Lmna−/− mice. The analysis included the percentage of positive cells and the membrane expression of MHC-II and co-stimulatory molecules, as well as the intracellular levels of IL-6 and TNFα, shown as intensity of positive cells. Data represent means ± SEM from at least 4 mice per group, analyzed using an unpaired Student’s t-test between genotypes. Statistical significance is denoted by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). This schematic was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution CC BY 4.0 License; https://smart.servier.com
Lamin A/C in BMDCs promotes nuclear localization of NF-κB
TLR4 signaling stimulates the expression of Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-related genes expression [53]. NF-κB plays a crucial role in regulating DC development, survival, and cytokine production [54]. As a master nuclear transcription factor for genes involved in inflammatory responses, RelA/p65, together with p50, constitutes the most common NF-κB subunits involved in the classical NF-κB pathway. NF-κB activity is tightly controlled by inhibitors, such as IκBα, which sequesters p65/p50 dimers in the cytoplasm. Activation of various signaling cascades leads to increased IκB kinase (IKK) activities, resulting in IκBα phosphorylation and subsequent degradation. This allows the release of homo- or hetero-dimers of NF-κB, enabling their translocation into the nucleus, where they activate gene transcription [55].
Given that previous studies have indicated lamin A/C role in enhancing the nuclear translocation of RelA/p65 and subsequently increasing NF-κB activity, leading to elevated inflammatory gene expression in macrophages [56], we sought to investigate whether a similar phenomenon occurs in our BMDC model. To explore this, WT- or Lmna−/−-BMDCs were either treated or left untreated with LPS for 24 h, and confocal images of lamin A/C and NF-κB staining were acquired. The nucleus was identified by DAPI staining (Fig. 5). The analysis of NF-κB subunit, RelA, translocation to the nucleus is a well-established method for determining NF-κB activation [56]. Upon LPS addition, nuclear levels and the nuclear/cytoplasmic ratio of NF-κB significantly increased in WT BMDCs, without affecting total cellular NF-κB levels. However, Lmna−/− BMDCs did not exhibit statistically significantly increased nuclear levels, cellular amounts, or nuclear/cytoplasmic ratio of NF-κB upon LPS stimulation (Fig. 5A, B-E). In the absence of LPS, no differences were noted between genotypes in total cellular NF-κB levels or the NF-κB nuclear/cytoplasmic ratio, except for reduced NF-κB nuclear localization in Lmna−/− BMDCs compared to WT BMDCs (Fig. 5B-E). In WT-BMDCs with LPS, RelA was predominantly in the nucleus, while in Lmna−/−-BMDCs with LPS, RelA was enriched in the cytoplasm, indicating significant differences in nuclear NF-κB levels and nuclear/cytoplasmic ratio (Fig. 5A). Additionally, there was a correlation (Pearson two-tailed, P < 0.0001) between lamin A/C and NF-κB nuclear levels in both LPS-unmatured and LPS-matured WT cells (Fig. 5F). These findings suggest that lamin A/C potentially promotes the nuclear presence of NF-κB in WT-BMDCs, highlighting its role in modulating NF-κB gene-dependent transcription.
Lamin A/C in GM-CSF-BMDCs favors NF-κB presence inside the nucleus. WT or Lmna−/− GM-CSF-BMDCs, either treated or untreated with LPS for 24 h, were stained with anti-lamin A/C and anti-NF-κB antibodies along with DAPI. (A) Representative micrographs of LPS-treated WT or Lmna−/− GM-CSF-BMDCs. Quantification of (B) lamin A/C levels, (C) nuclear NF-κB levels, (D) cellular NF-κB levels, and (E) nuclear/cytoplasmic NF-κB ratio. Data are presented as means ± SEM from two independently conducted experiments and were analyzed by one-way ANOVA with Bonferroni’s multiple comparison test (*P < 0.05; ***P < 0.001; ns = not significant; A.U.=Arbitrary units). (F) Correlation between nuclear NF-κB levels and lamin A/C levels in WT GM-CSF-BMDCs, either treated or untreated with LPS. Each symbol in the graphs represents an individual cell (n ≥ 20). The analysis was conducted using linear regression and correlation. This schematic was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution CC BY 4.0 License; https://smart.servier.com
Lamin A/C in BMDCs enhances the expression of NF-κB-related genes
As lamin A/C in BMDCs appeared to promote the presence of NF-κB in the nucleus, we conducted qPCR analysis of genes whose transcription is modulated by this transcription factor. mRNA from WT- and Lmna−/−-BMDCs stimulated with LPS for 24 h was extracted, and NF-κB-related gene expression patterns were validated by RT-qPCR (Fig. 6). Lmna−/−-BMDCs expressed lower levels of the mRNA of proinflammatory molecules such as IL-1α, IL-2, or IFNβ, as well as the anti-inflammatory cytokine IL-10. NF-κB regulates the transcription of genes involved in other functions, including Ccr7 associated with DC migration towards LN, Cxcr2 important for maintaining the fate of normal hematopoietic stem/progenitor cells, Icam1 that encodes an adhesion molecule, and Mip1α, a crucial molecule produced by macrophages and monocytes after bacterial endotoxin stimulation. Expression of these genes was statistically reduced in Lmna−/−-BMDCs compared to WT cells.
NF-κB-dependent gene expression profiling in WT and Lmna−/−-GM-CSF BMDCs stimulated with LPS for 24 h. mRNA expression levels of NF-κB-dependent genes in WT- and Lmna−/−-BMDCs. The analysis involved RT-qPCR of specified NF-κB-dependent genes in BMDCs stimulated with LPS for 24 h (n = 3, from 3 independent experiments). The presented data represents means ± SEM and were analyzed by unpaired Student’s t-test. Significance levels are indicated by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). Housekeeping genes Ywhaz, β-actin, and Rna18s1 were used for normalization. This schematic was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution CC BY 4.0 License; https://smart.servier.com
These results suggest that lamin A/C may regulate the production of various pro-inflammatory cytokines and other immune molecules, possibly through the modulation of NF-κB localization, among other processes.
Lamin A/C shapes chromatin accessibility in BMDCs, modulating antiviral response
Lamin A/C is a well-known epigenetic regulator [29, 57], yet its influence on transcriptional modulation in DCs during pathogenic infections remains elusive. We aimed to investigate whether alterations in the gene accessibility profile of Lmna−/−-BMDCs in response to LPS stimulation show a distinct innate phenotype compared to WT-BMDCs. To this end, we employed ATAC-seq, a technique that allows for the examination of changes in chromatin accessibility. Isolated nuclei from LPS-treated WT- and Lmna−/−-BMDCs were treated with transposase 5 (Tn5) to identify regions undergoing active chromatin remodeling. Overall, we found that Lmna-/--BMDCs have more closed chromatin, as indicated by the lower blue profile near Transcription Start Site (TSS) (Fig. 7A). Our analysis identified approximately 21,000 well defined ATAC-peaks, 2,884 of which exhibited differential accessibility between conditions (corrected pValue < 0.05) (Fig. 7B). Of these, 2,840 had lower coverage in Lmna−/−-BMDCs compared to WT. As expected, the majority of differential peaks were located in intronic or intergenic regions, spanning 1 to 100 kb from transcription start sites, indicating their association with regulatory regions. Among the 2,840 closed peaks in Lmna−/−-BMDCs, 2,152 (75%) were in promoters, 197 (6.9%) in CpG Islands, and 177 (6.2%) in 5’ UTR regions, highlighting their importance in transcription regulation. In contrast, of the 44 wider peaks in Lmna−/−-BMDCs only 10 (22.7%) were in regulatory regions, suggesting they are less significant at the transcriptional level.
Differential gene accessibility signature in WT and Lmna−/− GM-CSF BMDCs upon LPS stimulation. (A) ATAC-Seq aggregate profile around all genes of the genome. Mean coverage of both replicates of each condition is calculated and collapsed from 2,000 bp before Transcription Star Site (TSS) and after Transcription Termination Site (TTS). (B) Volcano plot of all well-defined ATAC peaks representing differential accessibility analysis. Differentially accessible peaks [with FDR < 0.05] are symbolized as colored dots, red ones for more accessible peaks in WT and blue-one for Lmna−/− BMDCs. (C) Over-Representation Analysis for genes with reduced accessibility in Lmna−/− BMDCs against Reactome pathway database colored by signification and ordered by gene ratio. (D) ATAC-Seq coverage profile for individual regions around promoter peak of Nfkbia, Pex2 and Txndc11 genes. Both replicas of WT (red) and Lmna−/− BMDCs (blue) are represented. Also, a mean profile for both replicas is represented overlapped for both conditions to enhance the difference. (E) mRNA expression levels measured by RT-qPCR analysis for the indicated genes in WT and Lmna−/− BMDCs stimulated with LPS for 24 h. The data, obtained from three independent experiments with three samples each, were analyzed using unpaired Student’s t-test. Housekeeping gene Rna18s1 was used for normalization. Data are presented as means ± SEM. (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). The schematic was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution CC BY 4.0 License; https://smart.servier.com
Functional enrichment analyses of genes associated with differentially accessible sites revealed biological processes consistent with DC and lamin A/C functionalities (Fig. 7C, Supplementary Fig. 2). Using the Reactome database as a reference (Fig. 7C), these biological pathways corresponded to established lamin A/C functions, such as gene expression [58], MAPK family signaling cascades, MAP kinase activation [59, 60], chromatin-modifying enzymes [28], and chromatin organization [61] (Fig. 7C). Additionally, other processes align with crucial functions associated with DCs in viral responses, including antigen processing or presentation, TLR signaling, including TLR4 and TLR2, as well as other cytokines (Fig. 7C) [62]. When we performed Over-Representation Analysis using other databases with functional information, such as Gene Ontology or Wikipathways (Supplementary Fig. 2A and C) we found similar results. Moreover, this secondary analysis confirmed that some of these processes directly align with the known functions of lamin A/C [24], including DNA repair, replication, and transcription, signaling and chromatin organization (Supplementary Fig. 2). Hence, these functional enrichment analyses suggest that lamin A/C in DCs, following LPS treatment, potentially regulates DCs active chromatin regions for a distinct antiviral response. When we examined molecular function enrichment (Supplementary Fig. 2B) we found more evidence of transcription regulation effects such as histone modification, RNA-binding, and epigenetic effectors. This data suggests reduced chromatin accessibility in Lmna−/−- BMDCs compared to WT-BMDCs in regions crucial for transcriptional regulation.
To validate the functionality of the accessibility of some of the regulatory regions of these genes, we confirmed that genes with lower chromatin accessibility in Lmna−/−-BMDCs, such as Tnxdc11, Pex2 and Nfkbia (Fig. 7B, D), were downregulated at the mRNA expression level (Fig. 7E). These differences in DNA accessibility between WT- and Lmna−/−-BMDCs may indicate a lamin A/C-dependent gene accessibility ability to modulate DC functions in the immune response against viruses.
Discussion
This study demonstrates that downregulation of lamin A/C in LysM-expressing myeloid cells suppresses their ability to generate effector CD8 CTL and Th1 responses in vivo, as well as a Th1 response in naïve T cells stimulated in vitro with BMDCs. This effect is mediated by changes in immune synapse formation, nuclear presence of NF-κB, and the gene accessibility signature, ultimately leading to the downregulation of proinflammatory gene expression.
The clearance of VACV in peripheral infection involves both CD8 CTL and Th1 cells through various mechanisms [35]. Approximately 7 days after VACV infection [63], there is an expansion of VACV-specific CD8 T cells peaking 10 days post-infection [41]. Additionally, the primary VACV-specific CD4 T cell response peaks at around 14 days post-inoculation, characterized by highly activated CD4 T cells producing IFNγ [64]. These IFNγ-producing CD4 T cells play a role not only as cytotoxic cells [65, 66] but also as helpers for CD8 CTLs, supporting the maintenance of the primary CD8 CTL response in an MHC-II-dependent manner [41].
In our study, there is a temporal correlation between the reduced VACV eradication observed at 14 days in Lmna−/− mice, reduced Th1 cell production at both 6 and 14 days, and decreased CD8 CTL production only at 14 days in lamin A/C-deficient mice upon s.s. These findings emphasize the importance of lamin A/C in the myeloid compartment for generating a Th1 and CD8 CTL response against the virus. They suggest that the increased viral accumulation in Lmna−/− mice at 14 days may result from reduced viral clearance due to the diminished CD8 CTL and Th1 response. It is not yet clear whether this reduced CD8 CTL response at 14 days is a direct result of the interaction with lamin A/C-deficient APCs or an indirect effect of the reduced Th1 response observed at 6 days, which may itself be a consequence of the absence of lamin A/C in APCs. Despite our results, the significance of CD4 T cells in the primary CD8 T cell response to VACV infection remains somewhat controversial [67], with some studies showing dependence [68,69,70], while others do not [22, 71, 72]. This variability in results may be linked to the route of infection and viral dose [41, 42]. Our results confirm that the route of VACV administration affects the primary CD4 and CD8 T cell responses since the absence of lamin A/C in APCs leads to differences in the CD8 CTL and Th1 responses depending on the administration route.
We were interested in establishing the cellular and molecular mechanisms that could be regulated by lamin A/C and thus modulate the functions of DCs in the antiviral response. Our results showed that lamin A/C absence in BMDCs stimulated with VACV lysate significantly reduced their ability to promote activation, proliferation, and differentiation into a Th1 phenotype. VACV can be recognized by TLR2 [45, 46] and TLR4 [47, 48] and indeed, the absence of lamin A/C in BMDCs matured with a TLR2 ligand inhibits their ability to promote activation and differentiation towards a Th1 phenotype, particularly at later times, without affecting early activation levels or T cell proliferation under the conditions studied. Additionally, after maturation of BMDCs with LPS, BMDCs have a reduced capacity to promote both early and late activation, as well as proliferation and differentiation into Th1 cells in co-cultures where cells lacking lamin A/C participate. This reduced effect observed after maturation of BMDCs with a TLR2 ligand, compared to the effects observed with VACV lysate or LPS, may be due to the greater stimulation of different receptors in the case of VACV and to a greater importance of lamin A/C in the pathways stimulated by LPS in the latter case. This led us to continue our research using LPS-mediated maturation of the TLR4 signaling pathway in the following experiments.
Given that lamin A/C regulates DC activity in Th1-type immune responses to viruses mediated by TLR4, it is likely that lamin A/C also modulates DC function in response to other pathogens recognized by TLR4, whether or not they elicit Th1 responses. For instance, TLR4 interaction with pathogen-associated molecular patterns (PAMPs) like O-mannan from fungi promote immune responses mediates by DC, macrophages or monocytes, including the production of pro-inflammatory cytokines essential for Th1 polarization. Such responses have been observed in vitro and in vivo to Cryptococcus neoformans [73], Candida albicans [74,75,76,77,78], and Aspergillus fumigatus [78,79,80,81]. TLR4 signaling is well-known for its role in recognizing LPS from Gram-negative bacteria, initiating inflammation, and often resulting in a Th1 response [82]. Responses to LPS via TLR4 have been described for bacteria such as Escherichia coli [83, 84], Porphyromonas gingivalis [83], Listeria monocytogenes [84], and Yersinia pestis [85]. Additionally, an alternative Th2 response has been associated to LPS from helmints [84].
Upon recognition of LPS by TLR4, DCs undergo a complex developmental program known as maturation, resulting in significant modifications in both structure and function [86]. This maturation enhances DC migration and upregulates the expression of costimulatory molecules, thereby increasing the capacity of DCs to present exogenous antigens [86].
In vivo, once matured and after incorporation, processing, and presentation on the membrane in the MHC-II context of VACV antigens, DCs migrate to a lymph node where they will specifically stimulate T cells, including CD4 T cells, through the formation of immune synapses [86]. The observed reduced capacity to generate Th1 cells could be related to a decreased formation of immune synapses by Lmna−/− DCs [26]. Our videomicroscopy experiments confirmed this possibility but without changes in MHC-II or costimulatory molecules.
In the mouse spleen, CD11c⁺ DCs comprise diverse subsets of DCs derived from bone marrow precursors distinct from monocytes [87, 88] including cDC1 (CD8α⁺ or CD103⁺ and CD11b+), which are proficient at cross-presenting antigens to CD8⁺ T cells, cDC2 (CD11b⁺, CD4⁺, MHC-II⁺), which specialize in presenting antigens to CD4⁺ T cells, and pDCs (B220⁺, PDCA-1⁺, Siglec-H⁺, Ly6C⁺), which are key producers of type I interferons in response to viral infections [89]. Additionally, monocyte-derived DCs, coming from blood, are present [90]. GM-CSF BMDCs stimulated with LPS exhibit similarities to in vivo dendritic cells in terms of activation markers (e.g., CD80, CD86), MHC class II expression, and secretion of pro-inflammatory cytokines like IL-12 and TNF-α [43, 91]. However, GM-CSF BMDCs, although derived from bone marrow precursors in vitro, more closely resemble inflammatory moDCs [92]. We tested whether certain populations of CD11c+ DCs in mouse spleen upon ex vivo LPS stimulation show similarities with LPS-GM-CSF BMDCs regarding the alterations observed due to the absence of lamin A/C. We found comparable alterations in proinflammatory cytokine production, in addition to reductions in MHC-II and some costimulatory molecules expression in membrane in CD11c+CD1b++ cells, a population with more monocyte-like profile. In contrast, CD11c++CD11b+ and CD11c++CD11b− cells, which may represent DCs with a non-monocytic origin in steady state, almost showed no alterations.
TLR4 signaling promotes NF-κB-related genes expression [53]. NF-κB plays a crucial role in regulating DC development, survival, and cytokine production [54]. Previous studies have indicated a role for lamin A/C in enhancing the nuclear translocation of RelA/p65 and subsequently increasing NF-κB activity, leading to elevated inflammatory gene expression in macrophages [56]. We observed that upon LPS addition, in WT-BMDCs, RelA was predominantly in the nucleus, while in Lmna−/−-BMDCs, RelA was enriched in the cytoplasm. These findings suggest that the absence of lamin A/C ameliorates the nuclear presence of NF-κB in the nucleus, potentially reducing NF-κB gene-dependent transcription.
The investigation into how lamin A/C triggers NF-κB translocation remains unresolved. Some researchers propose that lamin A/C activates IKKβ, leading to the phosphorylation and subsequent reduction of IκBα protein levels, thereby facilitating NF-κB nuclear translocation and transcriptional activation [56]. Another hypothesis suggests that the accumulation of prelamin A at the nuclear lamina activates the NF-κB pathway via NEMO-dependent signaling or inflammasome formation [93, 94]. Additionally, lamin A/C may modulate actin dynamics, influencing the nuclear translocation and downstream signaling of transcription factors like megakaryoblastic leukemia 1 (MLK1) [95]. Conversely, downregulation of lamin A/C could potentially diminish Rel A retention in the nucleus, resulting in NF-κB hypoactivation and the attenuation of inflammatory cascades. Another potential mechanism involves the regulation of nuclear NF-κB transcriptional activity through chromatin structure reorganization. Recent studies have illustrated changes in 3D genome organization during immune activation, wherein immune gene loci are released from the nuclear periphery to facilitate proper immune response activation [96]. Reduced lamin A/C levels may alter 3D chromatin organization or the interaction of the genome with the nuclear periphery, leading to dysregulated expression of inflammatory genes. To further explore this possibility, we investigated whether the absence of lamin A/C in DCs could modify the chromatin accessibility profile of these cells. Lamin A/C is a well-known epigenetic regulator [57], yet its impact on transcription modulation in DCs during pathogen infections remains unclear. We aimed to investigate whether alterations in the transcriptomic profile of Lmna−/−- BMDCs indicate a distinct innate phenotype in response to LPS stimulation compared to LPS-treated WT BMDCs. Using ATAC-seq, we assessed changes in chromatin accessibility. Functional enrichment analyses of genes associated with differentially accessible sites align directly with the known functions of lamin A/C in other contexts [24], and suggest the importance of lamin A/C in critical DC functions during viral infection. These findings expand our understanding of the role of lamin A/C in immune system cells, indicating that lamin A/C in DCs, following LPS treatment, primes DCs for an enhanced antiviral response.
ATAC-seq results also showed that the majority of differential peaks were situated in regulatory regions. Almost all peaks with significative differences (corrected pValue < 0.05) between WT and KO were wider in WT compared to Lmna−/− samples. This suggests reduced chromatin accessibility in Lmna−/−- BMDCs compared to WT BMDCs in regions crucial for transcriptional regulation, consistent with the increased general capacity for activation of T cells observed in previous figures.
In summary, these results underscore the critical role of lamin A/C in APCs, particularly moDCs and BMDCs, during the immune response against viral infections. They emphasize the importance of lamin A/C in optimal T cell activation, differentiation, and antiviral immune responses, providing insights into potential therapeutic strategies targeting APCs in infectious diseases.
Data availability
The data underlying this article are available in the article, in its online supplementary material, and in GEO under accession number GSE269613.
References
Swain SL, McKinstry KK, Strutt TM (2012) Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol 12(2):136–148. https://doi.org/10.1038/nri3152
Stewart AJ, Devlin PM (2006) The history of the smallpox vaccine. J Infect 52(5):329–334. https://doi.org/10.1016/j.jinf.2005.07.021
Norbury CC, Malide D, Gibbs JS, Bennink JR, Yewdell JW (2002) Visualizing priming of virus-specific CD8 + T cells by infected dendritic cells in vivo. Nat Immunol 3(3):265–271. https://doi.org/10.1038/ni762
Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K (2000) Immunobiology of dendritic cells. Annu Rev Immunol 18:767–811. https://doi.org/10.1146/annurev.immunol.18.1.767
Steinman RM (2012) Decisions about dendritic cells: past, present, and future. Annu Rev Immunol 30:1–22. https://doi.org/10.1146/annurev-immunol-100311-102839
Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, Shakhar G, Halpern Z, Jung S (2012) Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37(6):1076–1090. https://doi.org/10.1016/j.immuni.2012.08.026
Tian T, Jin MQ, Dubin K, King SL, Hoetzenecker W, Murphy GF, Chen CA, Kupper TS, Fuhlbrigge RC (2017) IL-1R type 1-Deficient mice demonstrate an impaired host Immune Response against Cutaneous Vaccinia Virus infection. J Immunol 198(11):4341–4351. https://doi.org/10.4049/jimmunol.1500106
Campbell IK, van Nieuwenhuijze A, Segura E, O’Donnell K, Coghill E, Hommel M, Gerondakis S, Villadangos JA, Wicks IP (2011) Differentiation of inflammatory dendritic cells is mediated by NF-kappaB1-dependent GM-CSF production in CD4 T cells. J Immunol 186(9):5468–5477. https://doi.org/10.4049/jimmunol.1002923
Steinman RM (2007) Dendritic cells: understanding immunogenicity. Eur J Immunol 37 Suppl 1S53–60. https://doi.org/10.1002/eji.200737400
Iwasaki A, Medzhitov R (2004) Toll-like receptor control of the adaptive immune responses. Nat Immunol 5(10):987–995. https://doi.org/10.1038/ni1112
Behzadi P, Garcia-Perdomo HA, Karpinski TM (2021) Toll-like receptors: General Molecular and Structural Biology. J Immunol Res 2021:9914854. https://doi.org/10.1155/2021/9914854
Wesa AK, Galy A (2001) IL-1 beta induces dendritic cells to produce IL-12. Int Immunol 13(8):1053–1061. https://doi.org/10.1093/intimm/13.8.1053
Medzhitov R (2001) Toll-like receptors and innate immunity. Nat Rev Immunol 1(2):135–145. https://doi.org/10.1038/35100529
Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J (2006) Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood 107(7):2613–2618. https://doi.org/10.1182/blood-2005-07-2965
Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA (1999) Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity 11(6):753 – 61 https://doi.org/10.1016/s1074-7613(00)80149-1
Trombetta ES, Mellman I (2005) Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol 23:975–1028. https://doi.org/10.1146/annurev.immunol.22.012703.104538
Trapani JA, Smyth MJ (2002) Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol 2(10):735–747. https://doi.org/10.1038/nri911
Sigal LJ (2016) The Pathogenesis and immunobiology of Mousepox. Adv Immunol 129:251–276. https://doi.org/10.1016/bs.ai.2015.10.001
Fang M, Siciliano NA, Hersperger AR, Roscoe F, Hu A, Ma X, Shamsedeen AR, Eisenlohr LC, Sigal LJ (2012) Perforin-dependent CD4 + T-cell cytotoxicity contributes to control a murine poxvirus infection. Proc Natl Acad Sci U S A 109(25):9983–9988. https://doi.org/10.1073/pnas.1202143109
Toribio-Fernandez R, Zorita V, Rocha-Perugini V, Iborra S, Martinez Del Hoyo G, Chevre R, Dorado B, Sancho D, Sanchez-Madrid F, Andres V, Gonzalez-Granado JM (2018) Lamin A/C augments Th1 differentiation and response against Vaccinia virus and Leishmania major. Cell Death Dis 9(1):9. https://doi.org/10.1038/s41419-017-0007-6
Fang M, Sigal LJ (2005) Antibodies and CD8 + T cells are complementary and essential for natural resistance to a highly lethal cytopathic virus. J Immunol 175(10):6829–6836. https://doi.org/10.4049/jimmunol.175.10.6829
Shedlock DJ, Shen H (2003) Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300(5617):337–339. https://doi.org/10.1126/science.1082305
Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP (2003) CD4 + T cells are required for secondary expansion and memory in CD8 + T lymphocytes. Nature 421(6925):852–856. https://doi.org/10.1038/nature01441
Andres V, Gonzalez JM (2009) Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol 187(7):945–957. https://doi.org/10.1083/jcb.200904124
Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ (2006) Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev 86(3):967–1008. https://doi.org/10.1152/physrev.00047.2005
Gonzalez-Granado JM, Silvestre-Roig C, Rocha-Perugini V, Trigueros-Motos L, Cibrian D, Morlino G, Blanco-Berrocal M, Osorio FG, Freije JMP, Lopez-Otin C, Sanchez-Madrid F, Andres V (2014) Nuclear envelope lamin-A couples actin dynamics with immunological synapse architecture and T cell activation. Sci Signal 7(322):ra37. https://doi.org/10.1126/scisignal.2004872
Rocha-Perugini V, Gonzalez-Granado JM (2014) Nuclear envelope lamin-A as a coordinator of T cell activation. Nucleus 5(5):396–401. https://doi.org/10.4161/nucl.36361
Toribio-Fernandez R, Herrero-Fernandez B, Zorita V, Lopez JA, Vazquez J, Criado G, Pablos JL, Collas P, Sanchez-Madrid F, Andres V, Gonzalez-Granado JM (2019) Lamin A/C deficiency in CD4(+) T-cells enhances regulatory T-cells and prevents inflammatory bowel disease. J Pathol 249(4):509–522. https://doi.org/10.1002/path.5332
Saez A, Herrero-Fernandez B, Gomez-Bris R, Somovilla-Crespo B, Rius C, Gonzalez-Granado JM (2020) Lamin A/C and the Immune System: one Intermediate Filament, many faces. Int J Mol Sci 21(17). https://doi.org/10.3390/ijms21176109
Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8(4):265–277. https://doi.org/10.1023/a:1008942828960
Kim Y, Zheng Y (2013) Generation and characterization of a conditional deletion allele for Lmna in mice. Biochem Biophys Res Commun 440(1):8–13. https://doi.org/10.1016/j.bbrc.2013.08.082
Rocha-Perugini V, Martinez Del Hoyo G, Gonzalez-Granado JM, Ramirez-Huesca M, Zorita V, Rubinstein E, Boucheix C, Sanchez-Madrid F (2017) CD9 regulates Major Histocompatibility Complex Class II trafficking in monocyte-derived dendritic cells. Mol Cell Biol 37(15). https://doi.org/10.1128/MCB.00202-17
Iborra S, Martinez-Lopez M, Khouili SC, Enamorado M, Cueto FJ, Conde-Garrosa R, Del Fresno C, Sancho D (2016) Optimal generation of tissue-Resident but not circulating memory T cells during viral infection requires crosspriming by DNGR-1(+) dendritic cells. Immunity 45(4):847–860. https://doi.org/10.1016/j.immuni.2016.08.019
Melamed S, Paran N, Katz L, Ben-Nathan D, Israely T, Schneider P, Levin R, Lustig S (2007) Tail scarification with Vaccinia virus Lister as a model for evaluation of smallpox vaccine potency in mice. Vaccine 25(45):7743–7753. https://doi.org/10.1016/j.vaccine.2007.09.023
Mota BE, Gallardo-Romero N, Trindade G, Keckler MS, Karem K, Carroll D, Campos MA, Vieira LQ, da Fonseca FG, Ferreira PC, Bonjardim CA, Damon IK, Kroon EG (2011) Adverse events post smallpox-vaccination: insights from tail scarification infection in mice with Vaccinia virus. PLoS ONE 6(4):e18924. https://doi.org/10.1371/journal.pone.0018924
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov (Camb) 2(3):100141. https://doi.org/10.1016/j.xinn.2021.100141
Yu G, He QY (2016) ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 12(2):477–479. https://doi.org/10.1039/c5mb00663e
De Silva NS, Siewiera J, Alkhoury C, Nader GPF, Nadalin F, de Azevedo K, Couty M, Izquierdo HM, Bhargava A, Conrad C, Maurin M, Antoniadou K, Fouillade C, Londono-Vallejo A, Behrendt R, Bertotti K, Serdjebi C, Lanthiez F, Gallwitz L, Saftig P, Herrero-Fernandez B, Saez A, Gonzalez-Granado JM, van Niel G, Boissonnas A, Piel M, Manel N (2023) Nuclear envelope disruption triggers hallmarks of aging in lung alveolar macrophages. Nat Aging 3(10):1251–1268. https://doi.org/10.1038/s43587-023-00488-w
Munier CML, van Bockel D, Bailey M, Ip S, Xu Y, Alcantara S, Liu SM, Denyer G, Kaplan W, group, Suzuki PS, Croft K, Purcell N, Tscharke A, Cooper D, Kent DA, Zaunders SJ, Kelleher JJ (2016) AD The primary immune response to Vaccinia virus vaccination includes cells with a distinct cytotoxic effector CD4 T-cell phenotype. Vaccine 34(44):5251–5261 https://doi.org/10.1016/j.vaccine.2016.09.009
Toribio-Fernandez R, Zorita V, Herrero-Fernandez B, Gonzalez-Granado JM (2018) An in vivo mouse model to measure naive CD4 T cell activation, proliferation and Th1 differentiation Induced by Bone Marrow-derived dendritic cells. J Vis Exp (138). https://doi.org/10.3791/58118
Hu Z, Molloy MJ, Usherwood EJ (2016) CD4(+) T-cell dependence of primary CD8(+) T-cell response against Vaccinia virus depends upon route of infection and viral dose. Cell Mol Immunol 13(1):82–93. https://doi.org/10.1038/cmi.2014.128
Pan Y, Liu L, Tian T, Zhao J, Park CO, Lofftus SY, Stingley CA, Yan Y, Mei S, Liu X, Kupper TS (2021) Epicutaneous immunization with modified vaccinia Ankara viral vectors generates superior T cell immunity against a respiratory viral challenge. NPJ Vaccines 6(1):1. https://doi.org/10.1038/s41541-020-00265-5
Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, Goubau D, Reis e Sousa C (2015) GM-CSF mouse bone marrow cultures comprise a Heterogeneous Population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity 42(6):1197–1211. https://doi.org/10.1016/j.immuni.2015.05.018
Barbalat R, Lau L, Locksley RM, Barton GM (2009) Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol 10(11):1200–1207. https://doi.org/10.1038/ni.1792
Zhu J, Martinez J, Huang X, Yang Y (2007) Innate immunity against Vaccinia virus is mediated by TLR2 and requires TLR-independent production of IFN-beta. Blood 109(2):619–625. https://doi.org/10.1182/blood-2006-06-027136
O’Gorman WE, Sampath P, Simonds EF, Sikorski R, O’Malley M, Krutzik PO, Chen H, Panchanathan V, Chaudhri G, Karupiah G, Lewis DB, Thorne SH, Nolan GP (2010) Alternate mechanisms of initial pattern recognition drive differential immune responses to related poxviruses. Cell Host Microbe 8(2):174–185. https://doi.org/10.1016/j.chom.2010.07.008
Lysakova-Devine T, Keogh B, Harrington B, Nagpal K, Halle A, Golenbock DT, Monie T, Bowie AG (2010) Viral inhibitory peptide of TLR4, a peptide derived from vaccinia protein A46, specifically inhibits TLR4 by directly targeting MyD88 adaptor-like and TRIF-related adaptor molecule. J Immunol 185(7):4261–4271. https://doi.org/10.4049/jimmunol.1002013
Kim Y, Lee H, Heo L, Seok C, Choe J (2014) Structure of Vaccinia virus A46, an inhibitor of TLR4 signaling pathway, shows the conformation of VIPER motif. Protein Sci 23(7):906–914. https://doi.org/10.1002/pro.2472
Davies ML, Sei JJ, Siciliano NA, Xu RH, Roscoe F, Sigal LJ, Eisenlohr LC, Norbury CC (2014) MyD88-dependent immunity to a natural model of Vaccinia virus infection does not involve toll-like receptor 2. J Virol 88(6):3557–3567. https://doi.org/10.1128/JVI.02776-13
Dillon S, Agrawal A, Van Dyke T, Landreth G, McCauley L, Koh A, Maliszewski C, Akira S, Pulendran B (2004) A toll-like receptor 2 ligand stimulates Th2 responses in vivo, via induction of extracellular signal-regulated kinase mitogen-activated protein kinase and c-Fos in dendritic cells. J Immunol 172(8):4733–4743. https://doi.org/10.4049/jimmunol.172.8.4733
Bustos-Moran E, Blas-Rus N, Martin-Cofreces NB, Sanchez-Madrid F (2016) Orchestrating lymphocyte polarity in Cognate Immune cell-cell interactions. Int Rev Cell Mol Biol 327:195–261. https://doi.org/10.1016/bs.ircmb.2016.06.004
Villadangos JA, Schnorrer P, Wilson NS (2005) Control of MHC class II antigen presentation in dendritic cells: a balance between creative and destructive forces. Immunol Rev 207:191–205. https://doi.org/10.1111/j.0105-2896.2005.00317.x
Kawai T, Akira S (2007) Signaling to NF-kappaB by toll-like receptors. Trends Mol Med 13(11):460–469. https://doi.org/10.1016/j.molmed.2007.09.002
Ouaaz F, Arron J, Zheng Y, Choi Y, Beg AA (2002) Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity 16(2):257–270. https://doi.org/10.1016/s1074-7613(02)00272-8
Scheidereit C (2006) IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene 25(51):6685–6705. https://doi.org/10.1038/sj.onc.1209934
Kim Y, Bayona PW, Kim M, Chang J, Hong S, Park Y, Budiman A, Kim YJ, Choi CY, Kim WS, Lee J, Cho KW (2018) Macrophage lamin A/C regulates inflammation and the development of obesity-Induced insulin resistance. Front Immunol 9:696. https://doi.org/10.3389/fimmu.2018.00696
Wang Y, Dobreva G (2023) Epigenetics in LMNA-Related cardiomyopathy. Cells 12(5). https://doi.org/10.3390/cells12050783
Ivorra C, Kubicek M, Gonzalez JM, Sanz-Gonzalez SM, Alvarez-Barrientos A, O’Connor JE, Burke B, Andres V (2006) A mechanism of AP-1 suppression through interaction of c-Fos with lamin A/C. Genes Dev 20(3):307–320. https://doi.org/10.1101/gad.349506
Gonzalez JM, Navarro-Puche A, Casar B, Crespo P, Andres V (2008) Fast regulation of AP-1 activity through interaction of lamin A/C, ERK1/2, and c-Fos at the nuclear envelope. J Cell Biol 183(4):653–666. https://doi.org/10.1083/jcb.200805049
Rodriguez J, Calvo F, Gonzalez JM, Casar B, Andres V, Crespo P (2010) ERK1/2 MAP kinases promote cell cycle entry by rapid, kinase-independent disruption of retinoblastoma-lamin A complexes. J Cell Biol 191(5):967–979. https://doi.org/10.1083/jcb.201004067
Briand N, Collas P (2018) Laminopathy-causing lamin A mutations reconfigure lamina-associated domains and local spatial chromatin conformation. Nucleus 9(1):216–226. https://doi.org/10.1080/19491034.2018.1449498
Carroll SL, Pasare C, Barton GM (2024) Control of adaptive immunity by pattern recognition receptors. Immunity 57(4):632–648. https://doi.org/10.1016/j.immuni.2024.03.014
Kaech SM, Wherry EJ (2007) Heterogeneity and cell-fate decisions in effector and memory CD8 + T cell differentiation during viral infection. Immunity 27(3):393–405. https://doi.org/10.1016/j.immuni.2007.08.007
Zaunders JJ, Dyer WB, Munier ML, Ip S, Liu J, Amyes E, Rawlinson W, De Rose R, Kent SJ, Sullivan JS, Cooper DA, Kelleher AD (2006) CD127 + CCR5 + CD38+++ CD4 + Th1 effector cells are an early component of the primary immune response to Vaccinia virus and precede development of interleukin-2 + memory CD4 + T cells. J Virol 80(20):10151–10161. https://doi.org/10.1128/JVI.02670-05
Mitra-Kaushik S, Cruz J, Stern LJ, Ennis FA, Terajima M (2007) Human cytotoxic CD4 + T cells recognize HLA-DR1-restricted epitopes on Vaccinia virus proteins A24R and D1R conserved among poxviruses. J Immunol 179(2):1303–1312. https://doi.org/10.4049/jimmunol.179.2.1303
Siciliano NA, Hersperger AR, Lacuanan AM, Xu RH, Sidney J, Sette A, Sigal LJ, Eisenlohr LC (2014) Impact of distinct poxvirus infections on the specificities and functionalities of CD4 + T cell responses. J Virol 88(17):10078–10091. https://doi.org/10.1128/JVI.01150-14
Wiesel M, Oxenius A (2012) From crucial to negligible: functional CD8(+) T-cell responses and their dependence on CD4(+) T-cell help. Eur J Immunol 42(5):1080–1088. https://doi.org/10.1002/eji.201142205
Wiesel M, Joller N, Ehlert AK, Crouse J, Sporri R, Bachmann MF, Oxenius A (2010) Th cells act via two synergistic pathways to promote antiviral CD8 + T cell responses. J Immunol 185(9):5188–5197. https://doi.org/10.4049/jimmunol.1001990
Obar JJ, Molloy MJ, Jellison ER, Stoklasek TA, Zhang W, Usherwood EJ, Lefrancois L (2010) CD4 + T cell regulation of CD25 expression controls development of short-lived effector CD8 + T cells in primary and secondary responses. Proc Natl Acad Sci U S A 107(1):193–198. https://doi.org/10.1073/pnas.0909945107
Novy P, Quigley M, Huang X, Yang Y (2007) CD4 T cells are required for CD8 T cell survival during both primary and memory recall responses. J Immunol 179(12):8243–8251. https://doi.org/10.4049/jimmunol.179.12.8243
Fuse S, Tsai CY, Molloy MJ, Allie SR, Zhang W, Yagita H, Usherwood EJ (2009) Recall responses by helpless memory CD8 + T cells are restricted by the up-regulation of PD-1. J Immunol 182(7):4244–4254. https://doi.org/10.4049/jimmunol.0802041
Fang M, Remakus S, Roscoe F, Ma X, Sigal LJ (2015) CD4 + T cell help is dispensable for protective CD8 + T cell memory against mousepox virus following Vaccinia virus immunization. J Virol 89(1):776–783. https://doi.org/10.1128/JVI.02176-14
Shoham S, Huang C, Chen JM, Golenbock DT, Levitz SM (2001) Toll-like receptor 4 mediates intracellular signaling without TNF-alpha release in response to Cryptococcus neoformans polysaccharide capsule. J Immunol 166(7):4620–4626. https://doi.org/10.4049/jimmunol.166.7.4620
Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ (2002) The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis 185(10):1483–1489. https://doi.org/10.1086/340511
Jouault T, Ibata-Ombetta S, Takeuchi O, Trinel PA, Sacchetti P, Lefebvre P, Akira S, Poulain D (2003) Candida albicans Phospholipomannan is sensed through toll-like receptors. J Infect Dis 188(1):165–172. https://doi.org/10.1086/375784
Blasi E, Mucci A, Neglia R, Pezzini F, Colombari B, Radzioch D, Cossarizza A, Lugli E, Volpini G, Del Giudice G, Peppoloni S (2005) Biological importance of the two toll-like receptors, TLR2 and TLR4, in macrophage response to infection with Candida albicans. FEMS Immunol Med Microbiol 44(1):69–79. https://doi.org/10.1016/j.femsim.2004.12.005
Bojang E, Ghuman H, Kumwenda P, Hall RA (2021) Immune Sensing of Candida albicans. J Fungi (Basel) 7(2). https://doi.org/10.3390/jof7020119
Bellocchio S, Montagnoli C, Bozza S, Gaziano R, Rossi G, Mambula SS, Vecchi A, Mantovani A, Levitz SM, Romani L (2004) The contribution of the Toll-like/IL-1 receptor superfamily to innate and adaptive immunity to fungal pathogens in vivo. J Immunol 172(5):3059–3069. https://doi.org/10.4049/jimmunol.172.5.3059
Wang JE, Warris A, Ellingsen EA, Jorgensen PF, Flo TH, Espevik T, Solberg R, Verweij PE, Aasen AO (2001) Involvement of CD14 and toll-like receptors in activation of human monocytes by aspergillus fumigatus hyphae. Infect Immun 69(4):2402–2406. https://doi.org/10.1128/IAI.69.4.2402-2406.2001
Mambula SS, Sau K, Henneke P, Golenbock DT, Levitz SM (2002) Toll-like receptor (TLR) signaling in response to Aspergillus Fumigatus. J Biol Chem 277(42):39320–39326. https://doi.org/10.1074/jbc.M201683200
Meier A, Kirschning CJ, Nikolaus T, Wagner H, Heesemann J, Ebel F (2003) Toll-like receptor (TLR) 2 and TLR4 are essential for aspergillus-induced activation of murine macrophages. Cell Microbiol 5(8):561–570. https://doi.org/10.1046/j.1462-5822.2003.00301.x
Re F, Strominger JL (2001) Toll-like receptor 2 (TLR2) and TLR4 differentially activate human dendritic cells. J Biol Chem 276(40):37692–37699. https://doi.org/10.1074/jbc.M105927200
Su H, Yan X, Dong Z, Chen W, Lin ZT, Hu QG (2015) Differential roles of Porphyromonas gingivalis lipopolysaccharide and Escherichia coli lipopolysaccharide in maturation and antigen-presenting functions of dentritic cells. Eur Rev Med Pharmacol Sci 19(13):2482–2492
van Riet E, Everts B, Retra K, Phylipsen M, van Hellemond JJ, Tielens AG, van der Kleij D, Hartgers FC, Yazdanbakhsh M (2009) Combined TLR2 and TLR4 ligation in the context of bacterial or helminth extracts in human monocyte derived dendritic cells: molecular correlates for Th1/Th2 polarization. BMC Immunol 10:9. https://doi.org/10.1186/1471-2172-10-9
Hajjar AM, Ernst RK, Fortuno ES 3rd, Brasfield AS, Yam CS, Newlon LA, Kollmann TR, Miller SI, Wilson CB (2012) Humanized TLR4/MD-2 mice reveal LPS recognition differentially impacts susceptibility to Yersinia pestis and Salmonella enterica. PLoS Pathog 8(10):e1002963. https://doi.org/10.1371/journal.ppat.1002963
Alloatti A, Kotsias F, Magalhaes JG, Amigorena S (2016) Dendritic cell maturation and cross-presentation: timing matters! Immunol Rev 272(1):97–108. https://doi.org/10.1111/imr.12432
Xu L, Kwak M, Zhang W, Lee PC, Jin JO (2017) Time-dependent effect of E. Coli LPS in spleen DC activation in vivo: alteration of numbers, expression of co-stimulatory molecules, production of pro-inflammatory cytokines, and presentation of antigens. Mol Immunol 85:205–213. https://doi.org/10.1016/j.molimm.2017.02.017
Shortman K, Liu YJ (2002) Mouse and human dendritic cell subtypes. Nat Rev Immunol 2(3):151–161. https://doi.org/10.1038/nri746
Pillarisetty VG, Shah AB, Miller G, Bleier JI, DeMatteo RP (2004) Liver dendritic cells are less immunogenic than spleen dendritic cells because of differences in subtype composition. J Immunol 172(2):1009–1017. https://doi.org/10.4049/jimmunol.172.2.1009
Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, Jeffrey KL, Anthony RM, Kluger C, Nchinda G, Koh H, Rodriguez A, Idoyaga J, Pack M, Velinzon K, Park CG, Steinman RM (2010) Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell 143(3):416–429. https://doi.org/10.1016/j.cell.2010.09.039
Na YR, Jung D, Gu GJ, Seok SH (2016) GM-CSF grown bone marrow derived cells are composed of phenotypically different dendritic cells and macrophages. Mol Cells 39(10):734–741. https://doi.org/10.14348/molcells.2016.0160
Zhang W, Ding Y, Sun L, Hong Q, Sun Y, Han L, Zi M, Xu Y (2018) Bone marrow-derived inflammatory and steady state DCs are different in both functions and survival. Cell Immunol 331:100–109. https://doi.org/10.1016/j.cellimm.2018.06.001
Osorio FG, Barcena C, Soria-Valles C, Ramsay AJ, de Carlos F, Cobo J, Fueyo A, Freije JM, Lopez-Otin C (2012) Nuclear lamina defects cause ATM-dependent NF-kappaB activation and link accelerated aging to a systemic inflammatory response. Genes Dev 26(20):2311–2324. https://doi.org/10.1101/gad.197954.112
Di Micco A, Frera G, Lugrin J, Jamilloux Y, Hsu ET, Tardivel A, De Gassart A, Zaffalon L, Bujisic B, Siegert S, Quadroni M, Broz P, Henry T, Hrycyna CA, Martinon F (2016) AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity. Proc Natl Acad Sci U S A 113(32):E4671–E4680. https://doi.org/10.1073/pnas.1602419113
Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J (2013) Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 497(7450):507–511. https://doi.org/10.1038/nature12105
Robson MI, de Las Heras JI, Czapiewski R, Sivakumar A, Kerr ARW, Schirmer EC (2017) Constrained release of lamina-associated enhancers and genes from the nuclear envelope during T-cell activation facilitates their association in chromosome compartments. Genome Res 27(7):1126–1138. https://doi.org/10.1101/gr.212308.116
Acknowledgements
We would like to thank Hector Sanchez Martinez and Tamara del Castillo for technical assistance.
Funding
This work was supported by grants from Ministerio de Ciencia, Innovación y Universidades (MCNU from Spain (grant number RTI2018-097504-B-I00; PID2021-125415OB-I00), Ministerio de Economía y Competitividad (grant number PID-2020-120412RB-I00) and La Caixa Health Research Grant (grant number LCF/PR/HR23/52430018). Instituto de Salud Carlos III (ISCIII) (grant number PI20/00306 and PI24/00146) with co-funding from the European Regional Development Fund (ERDF) “A way to build Europe”. The CNIC is supported by the ISCIII, the Ministerio de Ciencia, Innovación y Universidades (MCNU), and the Pro CNIC Foundation. B.H.-F. and R.G.-B. by the UAM and the MCNU FPU program (FPU18/00895, FPU19/01774); A.S. by Universidad Francisco de Vitoria; and H.S-M. by the Comunidad de Madrid YEI program (PEJ-2020-TL/BMD-17604).
Author information
Authors and Affiliations
Contributions
B.H.F; R.G.B; M.O.Z: Data curation, Formal Analysis, Investigation, Methodology. Software, Validation, Visualization. S.I; F.S.M: Conceptualization, Funding acquisition, Investigation, Project administration, Writing-review & editing. A.Q; A.D: Funding acquisition, review & editing. E.V: Data curation, Formal Analysis, Investigation, Writing-review & editing. A.S: Conceptualization, Formal Analysis, Investigation, Methodology, Writing-original draft, Writing-review & editing. V.Z: Data curation, Formal Analysis, Investigation, Methodology. S.M.A: Conceptualization, Investigation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Visualization, Writing-review & editing. J.M.G.G. Conceptualization, Data curation, Formal Analysis, Investigation, Funding acquisition, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing-original draft, Writing-review & editing.
Corresponding authors
Ethics declarations
Ethical approval
Experimental procedures were approved by the Animal Care and Ethics Committee of the CNIC, UAM, and the regional authorities approved all experimental procedures.
Consent for publication
Not applicable.
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Herrero-Fernández, B., Ortega-Zapero, M., Gómez-Bris, R. et al. Role of lamin A/C on dendritic cell function in antiviral immunity. Cell. Mol. Life Sci. 81, 400 (2024). https://doi.org/10.1007/s00018-024-05423-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05423-9