Abstract
Although the Hepatitis E virus (HEV) is an emerging global health burden, little is known about its interaction with the host cell. HEV genome encodes three proteins including the ORF2 capsid protein that is produced in different forms, the ORF2i protein which is the structural component of viral particles, and the ORF2g/c proteins which are massively secreted but are not associated with infectious material. We recently demonstrated that the endocytic recycling compartment (ERC) is hijacked by HEV to serve as a viral factory. However, host determinants involved in the subcellular shuttling of viral proteins to viral factories are unknown. Here, we demonstrate that the AP-1 adaptor complex plays a pivotal role in the targeting of ORF2i protein to viral factories. This complex belongs to the family of adaptor proteins that are involved in vesicular transport between the trans-Golgi network and early/recycling endosomes. An interplay between the AP-1 complex and viral protein(s) has been described for several viral lifecycles. In the present study, we demonstrated that the ORF2i protein colocalizes and interacts with the AP-1 adaptor complex in HEV-producing or infected cells. We showed that silencing or drug-inhibition of the AP-1 complex prevents ORF2i protein localization in viral factories and reduces viral production in hepatocytes. Modeling of the ORF2i/AP-1 complex also revealed that the S domain of ORF2i likely interacts with the σ1 subunit of AP-1 complex. Hence, our study identified for the first time a host factor involved in addressing HEV proteins (i.e. ORF2i protein) to viral factories.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis E virus (HEV) is a positive-strand RNA virus classified in the Paslahepevirus genus within the Hepeviridae family [1]. It represents the most common cause of acute viral hepatitis worldwide (WHO, 2022). Human individuals can be infected by five distinct genotypes (gt), belonging to a single serotype. HEV gt1 and gt2 are restricted to humans and are responsible for waterborne outbreaks in resource limited settings. HEV gt3, gt4, gt7 mainly cause a foodborne zoonosis due to consumption of undercooked meat foodstuffs in industrialized countries [2, 3]. Whilst hepatitis E infection predominantly triggers an asymptomatic and self-limiting disease, gt1 infection can lead to fulminant liver failure, especially in pregnant women. Infection with gt3, gt4 and gt7 can cause chronic disease in immunosuppressed patients. Moreover, chronic and acute HEV infections are associated with extrahepatic manifestations such as neurological disorders, kidney injuries or impaired renal function [4, 5]. Currently, there is no specific treatment or universal vaccine available.
HEV genome encodes three proteins namely ORF1, ORF2 and ORF3 proteins. ORF1 is a non-structural polyprotein that is essential for viral replication. ORF2 is the viral capsid protein. ORF3 is a protein involved in particle egress [6]. Previously, we and others demonstrated that during the HEV lifecycle, at least three forms of the ORF2 capsid protein are produced [7, 8] : (i) the non-infectious glycosylated ORF2g form (also named ORF2s [8]) is produced in large amounts in cell culture supernatant and is the most abundant antigen in patient sera [7, 9], (ii) the ORF2c form, derived from ORF2g cleavage [10], is found in patient sera and cell culture supernatant [7]. Both ORF2g and ORF2c forms (herein designated as ORF2g/c) presumably act as humoral decoys inhibiting antibody-mediated neutralization [8], (iii) the infectious ORF2i (also named ORF2c [8]) is the structural component of infectious particles that are likely derived from intracellular ORF2i assembly. Very recently, we showed that a short arginine-rich motif (ARM) in the ORF2 protein sequence regulates the fate and functions of HEV capsid protein forms. Thus, the ARM regulates the dual topology and functionality of the ORF2 signal peptide, leading to the production of either secreted non-infectious glycosylated ORF2g/c forms or cytosolic infectious ORF2i form. According to the positive inside rule in which positively charged amino acids (e.g., lysine, arginine) are more likely to be found on the cytoplasmic side of the membrane [11, 12], the ARM likely leads the ORF2i protein to be anchored by its N-terminus part (Nter) in the canonical secretion pathway membranes with its C-terminus (Cter) facing the cytosol. Thus, the ARM promotes ORF2-host cell membrane interactions [13]. More recently, we generated antibodies that specifically recognize the ORF2i protein, including the P1H1 antibody [9]. Thanks to these tools, we unveiled for the first time a membrane network composed of vesicular and tubular structures as HEV viral factories (i.e. endosomal recycling compartment or ERC) in perinuclear regions of HEV-producing cells. We demonstrated that these structures are enriched in ORF1, ORF2i, ORF3 proteins but also in viral RNA and ERC resident proteins (i.e. CD71 and Rab11). We showed that these structures are dependent on ORF2i protein assembly and the expression of ORF3 protein, which is a membrane-associated protein with palmitoylated N-terminal cysteine residues [14]. We also demonstrated that silencing the small GTPase Rab11 significatively impairs viral assembly and hereby viral infectivity without affecting viral replication [9].
Adaptors (or AP) are tetrameric complexes involved in the intracellular shuttling of membrane-bound proteins. Each complex is composed of two large subunits (a combination of γ/α/δ/ε/ς and β1–5, respectively), one medium subunit (µ1–5) and one small subunit (σ1–5) [15]. Five AP have been identified in humans [16, 17]. AP-1 and AP-2 mediate cargo transport in a clathrin-dependent manner, whereas AP-3, AP-4 and AP-5 are clathrin-independent [18]. The AP-1 complex is composed of γ and β1 large subunits, a µ1 medium subunit and a σ1 small subunit. It is involved in vesicular transport between the secretion pathway membranes (i.e. Trans-Golgi Network, TGN) and early/recycling endosomes [15, 19]. AP-1 complex subcellular localization relies on direct interaction with Arf1, a small guanine nucleotide-binding factor, and PI4P (Phosphatidylinositol-4-phosphate). Thus, AP-1 complex is recruited from the cytosol by its cargo, allowing its binding to clathrin and the formation of transport vesicles [20,21,22]. Several viruses such as SARS-CoV-2, Varicella-Zoster virus or Dengue virus directly or indirectly exploit AP-1 complex to promote viral infectivity [23,24,25].
Recent progress has been made in the understanding of the HEV lifecycle. However, cellular and/or viral determinants involved in the subcellular trafficking of viral components remain poorly characterized. In particular, addressing mechanisms of viral proteins, notably the ORF2i protein, to viral factories are unknown. In our study, by silencing the AP-1γ1 adaptin and by using a pharmacological inhibitor of AP-1 complex, we analyzed the involvement of the AP-1 complex in subcellular trafficking of the HEV ORF2i protein and its localization in viral factories. Our results highlight the importance of AP-1 complex-dependent shuttling for ORF2i protein addressing to viral factories and for the production of infectious viral particles in both hepatoma and primary cells. Molecular modeling also showed that ORF2i protein likely interacts with the σ1 subunit of AP-1 complex. Our study thus unveils for the first time a host factor involved in addressing the HEV ORF2i protein to viral factories.
Material & methods
Cell culture
PLC3, a subclone of PLC/PRF/5 hepatoma cells, and Huh-7.5 cells were cultured as previously described [10]. Both cell lines were authenticated by STR profiling and Multiplex Cell Authentication (Multiplexion), respectively. H7-T7-IZ cells, derived from Huh-7 cells and stably expressing the T7 RNA polymerase (kindly provided by R. Bartenschlager), were cultured in a medium supplemented with 50 µg/mL of Zeocin (Invivogen). They were transfected with the T7 promoter-driven pTM expression plasmids. PLC3 cells harboring a p6 subgenomic replicon were generated and cultured as previously described [9].
Plasmids, capped-RNA preparation and transfection
The plasmid pBlueScript SK(+) carrying the DNA of the full-length gt3 Kernow C-1 p6 strain (GenBank accession number JQ679013, kindly provided by S.U Emerson) was used. Capped genomic HEV RNAs were prepared with the mMESSAGE mMACHINE kit (Invitrogen) and delivered to PLC3 or Huh-7.5 cells by electroporation using a Gene Pulser Xcell apparatus (Bio-Rad). The cells were electroporated with p6 RNA (20 µg/4.106 cells). Mock PLC3 and Huh-7.5 cells were electroporated in the absence of RNA.
The pTM-ORF2-p6 (kindly provided by J. Gouttenoire, [26] and pTM-ORF2-Sar55 [13] plasmids were transfected into H7-T7-IZ cells using ViaFect (Promega) following the manufacturer’s recommendations.
Chemicals and viability assay
A5-membrane traffic inhibitor (A5, Calbiochem), Sofosbuvir (Sof, Selleckchem) and Ribavirin (RBV, MedChemExpress) were dissolved in water (A5) or DMSO (Sof and RBV), to generate a 50mM stock. Dose-response curve of PLC3 cells treated with A5 drug was performed. Cell viability was assessed using a CellTiter 96AQueous Non-Radioactive Cell Proliferation Assay (Promega) following the manufacturer’s instructions.
Silencing experiments
At 6 days post-electroporation (d.p.e.), PLC3/HEV cells were transfected with 20nM of small interfering RNA (siRNA) pools (Horizon) targeting the adaptin AP-1γ1 (ON-TARGETplus human AP1G1, gene 164, siRNA SMART-pool), the adaptin AP-1γ2 (ON-TARGETplus human AP1G2, gene 8906, siRNA SMART-pool), or with a nontargeting siRNA control (siCTL, ON-TARGETplus Non-targeting pool, siRNA SMART-pool) by using Lipofectamine RNAiMax reagent (Invitrogen) according to the manufacturer’s instructions. The knockdown effects were determined at 72 h post-transfection by western-blotting, immunoprecipitation, immunofluorescence, RNA and infectious titers measurements.
Antibodies
Primary antibodies used in this study are listed in Table 1. Peroxidase- and fluorochrome-conjugated secondary antibodies were obtained from Jackson ImmunoResearch.
Indirect immunofluorescence
Cells were grown on coverslips in 24-well plates and fixed at the indicated time points, with 3% of paraformaldehyde (PFA). After 20 min (min), cells were washed twice with phosphate-buffered saline (PBS) and incubated, or not, for 5 min with cold methanol. Then cells were permeabilized with 0.5% Triton X-100 (TX) for 30 min. Cells were incubated in PBS containing 10% goat serum for 30 min at room temperature (RT) and stained with the indicated primary antibodies for 30 min at RT followed by fluorochrome-conjugated secondary antibodies for 20 min at RT. The nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole, Invitrogen). After 2 washes with PBS, coverslips were mounted with Mowiol 4–88 (Calbiochem) on glass slides and analyzed with a LSM 880 confocal laser-scanning microscope (Zeiss) using a Plan Apochromat 63xOil/1.4 N.A. objective. The images were processed using Fiji software. The images shown in the figures are single confocal images.
Mander’s overlap coefficient (MOC) determination
Colocalization studies were performed by calculating the MOC using the JACoP plugin of Fiji software. For each analysis, at least 30 cells were selected to calculate a MOC mean. A MOC of 1 indicated a perfect overlap and 0 no overlap.
Western-blotting experiments
Samples were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (Hybond-ECL, Amersham). The targeted proteins were detected with specific antibodies that are described above and corresponding peroxidase-conjugated secondary antibodies. The detection of proteins was done by chemiluminescence analysis (ECL, Amersham).
Immunoprecipitations
For ORF2 protein immunoprecipitation experiments, mouse anti-ORF2 P1H1 and P3H2 monoclonal antibodies (mAb) [9] were bound to Epoxy Dynabeads™ M-270 (Invitrogen) overnight at 37 °C following the manufacturer’s recommendations. Irrelevant mouse IgG antibody (sc-2025, Santa Cruz Biotechnology) was used as a control. Beads were washed and then incubated for 1 h at room temperature with heat-denatured supernatants (20 min at 80 °C). Beads were washed and then heated at 80 °C for 20 min in Laemmli buffer. ORF2 proteins were detected by WB using the anti-ORF2 1E6 mAb antibody.
For co-immunoprecipitation experiments, rabbit anti-AP-1γ1 polyclonal antibody (pAb) or irrelevant anti-rabbit IgG (AB-105-C, Novus) were bound to Pierce™ magnetic A/G beads (Invitrogen). Cells were subjected to cross-linking using 0.8% PFA before lysis. Cell lysates were precleared on beads during 1 h at 4 °C. The precleared supernatant was added to beads-Ig complexes and incubated for 2 h at 4 °C. After the reaction, beads were washed six times with PBS 1X NP40 0.5% before elution with Laemmli buffer and boiled for 5 min at 95 °C. Samples were analyzed by Western blotting and inputs were 1/50 of the lysate volume subjected to co-immunoprecipitation. Rabbit anti-AP-1γ1 pAb and mouse anti-ORF2 1E6 were used for revelation.
Proximity ligation assay
Electroporated PLC3 cells cultured on glass coverslips were fixed with 3% paraformaldehyde for 20 min, methanol for 5 min and permeabilized in PBS containing 0.1% Triton X-100 for 30 min. Proximity ligation assay was performed using Duolink in situ detection kit (Sigma), as recommended by the manufacturer, with the anti-ORF2i P1H1 mAb and a polyclonal rabbit anti-AP-1γ1 antibody. Images were acquired by confocal microscopy. For each condition, twelve fields were acquired. For each field, a stack of images corresponding to the total volume of the cells was acquired. Maximum-intensity projection images were generated using Zen software. Representative images were assembled and dots were counted using Fiji software. For each field, the mean number of spots per cell was calculated by dividing the total number of spots by the total number of nuclei.
Infectious titer determination
Huh 7.5 cells were seeded in 96-well plates. The following day, cells were infected with serial dilutions of supernatants or intracellular viral particles from PLC3/HEV cells. Three days post-infection, cells were fixed and processed for indirect immunofluorescence. Cells labeled with anti-ORF2 1E6 mAb were counted as infected cells. The number of infected cells was determined for each dilution and used to define the infectious titers in focus forming unit/ml. Titers were adjusted to 100% for siCTL-transfected (siCTL) or diluent-treated (H2O or DMSO) cells.
Extraction of intracellular particles
Confluent T75 flasks of PLC3/HEV cells were trypsinized, and cells were centrifuged at 355g for 10 min. Cells were washed thrice with PBS. Intracellular viral particles were extracted by resuspending cells in 1 ml of sterile water at room temperature. Cells were vortexed vigorously for 20 min, and then, 110 µl of sterile 10X PBS was added. Samples were clarified by centrifugation at 19,100g for 10 min. The supernatants containing intracellular particles were collected and stored at − 80 °C until use.
RNA extraction and quantification
Extracellular and intracellular RNAs were extracted using QIAmp viral RNA mini kit (Qiagen) and Nucleospin RNA Plus kit (Macherey & Nagel), respectively. Next, extracted RNAs were converted to cDNA by using a polydT primer and the AffinityScript Multiple Temperature cDNA Synthesis kit (Agilent Technologies) according the manufacturer’s instructions. qPCR (TaqMan Gene Expression Assay, MGB-FAM-dye, ThermoFisher Scientific) was performed by using the QuantStudio3 Thermocycler (Applied Biosystems). For each sample, the same amounts of RNA were retro-transcribed and 1/4 of cDNA were used for qPCR. Intracellular and extracellular RNA levels were next calculated for 1 µg of RNA or 1 ml of supernatant, respectively. For quantification in PLC3/HEV cells, we used primers (5ʹ-GGTGGTTTCTGGGGTGAC-3ʹ (F) and 5ʹ-AGGGGTTGGTTG GATGAA-3ʹ (R)) and a probe (5ʹ-FAM-TGATTCTCAGCC CTTCGC-TAMRA-3ʹ) that target a conserved 70-bp region in the ORF2/3 overlap [27]. For cells harboring the subgenomic p6-replicon, quantifications were performed using primers (5’-AAGACATTCTGCGCTTTGTT-3’ (F) and 5’-TGACTCCTCATAAGCATCGC-3’ (R)) and a probe (5’-FAM-CCGTGGTTCCGTGCCATTGA-3’) that target the ORF1 [8].
Primary human hepatocytes (PHHs) experiments
Cryopreserved PHHs were obtained from Biopredic. PHHs (0,76 × 106 cells/well) were seeded in collagen I-coated 12-well plates in Williams E GlutaMAXTM-1 medium supplemented with 100 UI/mL penicillin, 100 µg/mL streptomycin, 4 µg/mL bovine insulin and 10% fetal calf serum v/v. One day post-seeding, PHHs were infected with HEV intracellular particles and treated with the corresponding drug or solvent in 500 µL of Williams E GlutaMAXTM-1 medium supplemented with 100 UI/mL penicillin, 100 µg/mL streptomycin, 4 µg/mL bovine insulin and 50 µM hydrocortisone. At D + 1 and D + 2, 250 µL of fresh medium containing drug/diluent were added to the wells. At D + 3, supernatants were recovered and processed for WB and extracellular infectious titers determination.
Modeling of putative ORF2i/AP-1 complexes and subcomplexes
We used an inhouse ColabFold (v. 1.5.2) [28] implementation of AlphaFold2 (v. 2.3) [29] to generate models of possible complexes of ORF2i with AP-1. Sequences for AP subunits were sp|Q10567 (human AP-1 complex subunit β1), sp|O43747 (human AP-1 complex subunit γ1), sp|Q9BXS5 (human AP-1 complex subunit µ1A), sp|Q9Y6Q5 (human AP-1 complex subunit µ1B), sp|P61966 (human AP-1 complex subunit σ1A, also referred to as σ1), sp|P56377 (human AP-1 complex subunit σ1B, also referred to as σ2), and sp|Q96PC3 (human AP-1 complex subunit σ1C, also referred to as σ3). For ORF2i, we used either the full-length sequence (residues 14–660 of the ORF2) or domain S only (residues 131–312). For sequence alignments the UniRef30 database was queried with MMseqs2 [30, 31]. These alignments were used as input to AlphaFold2 and 5 models generated for each. Resulting models were aligned to the S domain of PDB 6LAT. The models were displayed and rendered with the PyMOL Molecular Graphics System (version 2.5).
Multiple sequence alignment
ORF2 amino acid sequence alignment from 56 HEV strains was performed with Clustal Omega tool from EMBL-EBI [32].
Statistical analyses
Statistical analyses were performed using GraphPad Prism 9.5.0 software.
Results
HEV ORF2i protein colocalizes and interacts with the AP-1 adaptor complex
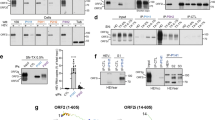
We have recently shown that the ERC is hijacked by HEV to serve as a viral factory [9]. Here, we attempted to identify the mechanisms underlying ORF2i localization to the ERC by analyzing the importance of the AP-1 complex in this process. First, PLC3 cells were electroporated with the infectious gt3 p6 strain RNA (PLC3/HEV) (Fig. 1a) or mock electroporated (PLC3) (Fig. S1), and the colocalization between HEV ORF2i protein (using P1H1 mAb that does not recognize the ORF2g/c forms) and the AP-1 adaptor complex was studied by confocal microscopy at 6 days post-electroporation (d.p.e). We analyzed the overlap of fluorescence intensities of ORF2i and AP-1 staining (Fig. 1a and Fig. S1) and calculated Manders’ overlap coefficients (MOC) of ORF2i protein staining in the AP-1 complex staining (Fig. 1f). As previously observed [9, 13], a nugget-like pattern was detected for the ORF2i protein in the perinuclear region. A perinuclear and dot-like staining was observed for the AP-1 complex (Fig. 1a and Fig. S1) which is in accordance with its TGN and endosomal localization. Interestingly, ORF2i protein and AP-1 complex staining highly overlapped in perinuclear regions (Fig. 1a, right panel), with a MOC value of 0.93 (Fig. 1f), indicating that both proteins likely colocalize in perinuclear regions of electroporated PLC3 cells. Importantly, confocal analyses of Huh-7.5 cells electroporated with p6-HEV RNA (Huh-7.5/HEV, Fig. 1b) or infected with HEV particles (Fig. 1c) also showed a colocalization of ORF2i protein and AP-1 complex in perinuclear regions with MOC values of 0.83 and 0.80, respectively (Fig. 1f). Super-resolution microscopy analysis also revealed a significant overlap of ORF2i protein/AP-1 complex fluorescence intensities in these contexts (Fig. 1b-c, right panels). These results indicate that ORF2i and AP-1 colocalization is not cell type-dependent and does not correspond to an artifact of electroporation.
HEV-gt1 and -gt3 ORF2i protein colocalizes with AP-1 complex in hepatoma cell lines. Electroporated PLC3/HEV (a) or Huh-7.5/HEV (b) cells were fixed at 6 d.p.e, while infected Huh-7.5 cells (c) were fixed at 12 d.p.i. The pTM-ORF2-p6 (d) and pTM-ORF2-Sar55 (e) transfected H7-T7-IZ cells were fixed at 24 h.p.t. Cells were then permeabilized with cold methanol and 0.5% Triton X-100 and double-stained with anti-ORF2i P1H1 and anti-AP-1γ1 antibodies. Staining were analyzed by confocal microscopy. Red = ORF2i; Green = AP-1; Blue = DAPI. Scale bar, 20 µM. (a-e) On the right, line graphs show overlaps of fluorescence intensities of ORF2i and AP-1 staining measured every 50 nm across the region of interest (ROI) highlighted by the white line on the “Merge” micrograph of each panel. (f) Mander’s overlap coefficients (MOC) of the ORF2i labelling in the AP-1 labelling using the whole cell as ROI. Each data dot in the bar chart represents a cell field, and the total number of ORF2-positive cells used to calculate the MOC is indicated. Displayed results come from biological replicates. Mann-Whitney test, ***p < 0.001, ****p < 0.0001
To strengthen our observations, we extended our study to HEV-gt1 by transfecting constructs expressing either HEV-gt1 ORF2 (Sar55 strain) or HEV-gt3 ORF2 (p6 strain) proteins in Huh-7 cells stably expressing the T7 RNA-polymerase (H7-T7-IZ cells) [33]. In this context, gt1 and gt3 ORF2i proteins display a similar subcellular localization pattern, as previously described [13]. They were characterized by a nugget-like perinuclear accumulation and peripheral distribution (Fig. 1d and e). Strikingly, both gt1 and gt3 ORF2i proteins highly colocalized with AP-1 complex in perinuclear regions with MOC values of 0.76 and 0.77 (Fig. 1f), respectively. In addition, as observed in electroporated/infected cells, peaks of fluorescence intensities of both proteins overlapped perfectly (Fig. 1d-e, right panels), indicating that the colocalization between ORF2i protein and AP-1 complex is conserved among HEV-gt1 and -gt3 strains.
AP-1γ1 adaptin silencing alters ORF2i protein localization in viral factories and inhibits viral replication and infectivity
We next evaluated the importance of the AP-1 complex in the HEV lifecycle by transfecting PLC3/HEV cells with small interfering RNA (siRNA) targeting the γ1 subunit of the AP-1 complex (siAP-1γ1) or non-targeting siRNA (siCTL) (Figs. 2 and 3). We silenced the AP-1γ1 subunit because many viruses, including Coronaviruses [24], subvert this adaptin to promote viral infectivity.
AP-1γ1 silencing affects ORF2i subcellular localization and colocalization with various viral/host cell markers. (a-d) At 6 d.p.e., PLC3/HEV cells were transfected with siRNA targeting AP-1γ1 (siAP-1γ1), with a non-targeting control siRNA (siCTL). At 3 d.p.t, cells were permeabilized with cold methanol and 0.5% Triton X-100 and double-stained with the indicated antibodies. For each double staining, MOC of the ORF2i labelling in the cellular marker/ORF3 labelling and MOC of the STX6 labelling in the TGN46 labelling were determined using the whole cell as ROI. Each data dot in the bar chart represents a cell field, and the total number of ORF2-positive cells used to calculate the MOC is indicated. Displayed results come from biological replicates. Mann-Whitney test, ****p < 0.0001
AP-1γ1 silencing affects viral RNA secretion and particle production. (a) Supernatants and cell lysates of non-transfected PLC3/HEV (PLC3/HEV/NT), PLC3/HEV/siCTL, PLC3/HEV/siAP-1γ1 or PLC3 cells were generated 3 days after siRNA transfection. In supernatants, ORF2i and ORF2g/c proteins were immunoprecipitated using anti-ORF2i P1H1 or anti-ORF2i/g/c P3H2 antibodies, respectively. An irrelevant mouse IgG antibody was used as an isotype control (Iso). ORF2 proteins were detected by WB using the 1E6 antibody. In cell lysates, silencing of AP-1γ1 was controlled by WB using a rabbit anti-AP-1γ1 antibody. ORF2i protein was detected using the 1E6 antibody. GRP78 and Tubulin proteins were detected using a rat anti-GRP78 antibody and a mouse anti-β-Tubulin antibody, respectively. (b) HEV RNA quantification in PLC3/HEV/siCTL, PLC3/HEV/siAP-1γ1, PLC3/HEV/DMSO, PLC3/HEV/Sofosbuvir-20µM or PLC3 cells after 3 days of transfection/treatment. Extracellular and intracellular viral RNAs were quantified by RT-qPCR. Titers were adjusted to 100% for siCTL/DMSO-treated cells. PLC3/HEV/Sofosbuvir-20µM cells were used as a positive control for replication inhibition. Values are from four independent experiments. Mann-Whitney test, ***p < 0.001, ****p < 0.0001. (c) Infectious titer determination in PLC3/HEV/siCTL, PLC3/HEV/siAP-1γ1, PLC3/HEV/DMSO, PLC3/HEV/Sofosbuvir-20µM or PLC3 cells after 3 days of transfection/treatment. Extracellular and intracellular viral particles were used to infect naïve Huh-7.5 cells for 3 days. Cells were next processed for indirect immunofluorescence. ORF2-positive cells were counted and each positive cell focus was considered as one FFU. Titers were adjusted to 100% for siCTL/DMSO-treated cells. PLC3/HEV/Sofosbuvir-20µM cells were used as a positive control for infectious titers inhibition. Values are from four independent experiments. Mann-Whitney test, **p < 0.01, ****p < 0.0001
We first carried out an extensive immunofluorescence analysis of the impact of AP-1γ1 silencing on HEV ORF2i protein subcellular localization and colocalization with viral/cellular markers, at 3 days post-transfection. Colocalization analyses were performed by using the P1H1 anti-ORF2i antibody and antibodies against markers related to AP-1 complex shuttling (AP-1, M6PR, Clathrin), TGN compartment (TGN46, STX6), ERC (Rab11) and HEV proteins (ORF3) (Fig. 2 and Fig. S2). Colocalization of markers was quantitatively analyzed by calculating the MOC (Fig. 2, right panels).
As expected, the ORF2i protein significantly colocalized with AP-1 complex in siCTL-transfected cells (MOC = 0.80) in perinuclear regions (Fig. 2a), whereas the ORF2i/AP-1 colocalization was abolished in siAP-1γ1-transfected cells (siAP-1γ1, MOC = 0.06). Interestingly, in AP-1γ1-knocked down cells, ORF2i protein displayed a more diffuse subcellular distribution as compared to siCTL-cells (Fig. 2a). Of note, we found that the siAP-1γ1 transfection had no impact on the subcellular localization of the AP-2 adaptor complex (Fig. S3a), which is involved in cargo endocytosis from plasma membrane to endosomes [15]. These results suggest that the AP-1 complex plays an important role in the subcellular addressing of ORF2i protein.
Next, we analyzed the colocalization between ORF2i protein and the cation-independent mannose-6-phosphate receptor (M6PR), a TGN resident protein that transits between TGN and endosomes [34, 35] (Fig. 2b). The AP-1 complex is known to bind and pack M6PR into transport vesicles at the TGN to deliver it as well as bound lysosomal enzymes to early endosomes, en route to late endosomes and lysosomes [36, 37]. When AP-1-dependent trafficking is disrupted, lysosomal enzymes are secreted instead of being addressed to lysosomes [38]. In PLC3/HEV cells transfected with siCTL, M6PR was detected in perinuclear regions, consistent with its TGN localization, and moderately colocalized with ORF2i protein (MOC = 0.44; Fig. 2b). In cells transfected with siAP-1γ1, M6PR displayed a diffuse dot-like staining in the cytosol, indicating an efficient inhibition of its AP-1-dependent shuttling, and a significant reduction of its colocalization with ORF2i protein was observed (MOC = 0.27; Fig. 2b). The ORF2i protein displayed a more diffuse staining in siAP-1γ1 cells where M6PR displayed a dot-like pattern, suggesting that silencing of AP-1 complex is likely responsible for altering its protein subcellular localization in PLC3/HEV cells.
To strengthen our observations, we examined the impact of AP-1 silencing on TGN compartment integrity to make certain that ORF2i subcellular localization modification was not due to a TGN morphological alteration in siRNA-transfected cells. For that purpose, we analyzed the colocalization of ORF2i protein with TGN46, (Fig. 2c) and colocalization of TGN46 with Syntaxin-6 (STX6), which are both markers of the TGN compartment [39, 40] (Fig. 2d). In siCTL and siAP-1γ1-transfected PLC3/HEV cells, TGN46 subcellular localization remained unchanged (Fig. 2c and d). Although weak, the codistribution of ORF2i with TGN46 was decreased significantly in siAP-1γ1 cells compared to siCTL cells (MOC = 0.18 vs. 0.32) (Fig. 2c). Importantly, subcellular localization and colocalization of TGN46 and STX6 remained unchanged upon siRNA transfection (Fig. 2d), indicating that the TGN integrity was not affected upon AP-1 silencing. Together these observations indicate that the AP-1 complex is likely involved in the subcellular trafficking of the HEV ORF2i protein.
Next, as AP-1 complex-dependent shuttling involves clathrin, we analyzed the colocalization between ORF2i protein and clathrin (Fig. 2e). In siCTL cells, a strong colocalization (MOC = 0.82) was observed in focalized perinuclear regions. In siAP-1γ1 cells, this colocalization was significantly reduced (MOC = 0.40) and clathrin-associated staining was more peripheral, indicating that AP-1 silencing also affects clathrin-recruitment to membranes. These results indicate that the ORF2i protein co-localizes with clathrin in an AP-1 complex-dependent manner.
We have previously shown that the ORF2i protein co-distributes in perinuclear nugget-like structures together with the HEV ORF3 protein and Rab11, a marker of ERC [9]. Therefore, we next studied the impact of AP-1 silencing on the colocalization of ORF2i with ORF3 (Fig. 2f) and Rab11 (Fig. 2g). Regarding the ORF2i/ORF3 co-staining, a diffuse cytosolic dot-like distribution of the HEV ORF3 protein was observed upon siAP-1γ1 transfection, as well as a significantly reduced ORF2i/ORF3 colocalization compared to control condition (MOC = 0.22 vs. 0.79; Fig. 2f). This indicates that AP-1 silencing alters the co-distribution of HEV ORF2i/ORF3 proteins. Regarding the ORF2i/Rab11 co-staining, a strong colocalization of both markers was observed in perinuclear nugget-like structures in control cells (MOC = 0.80; Fig. 2g) whereas their colocalization was significantly impaired upon siAP-1γ1 transfection (MOC = 0.37). These results indicate that the silencing of AP-1γ1 adaptin prevents the ORF2i protein to be targeted to ERC, and hence, to viral factories.
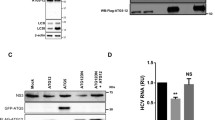
Next, we sought to assess the impact of altered ORF2i protein localization in viral factories upon AP-1γ1 on the viral lifecycle. For this purpose, we analyzed by Western blotting (WB) and immunoprecipitation (IP) the intracellular and extracellular ORF2 protein expression in PLC3/HEV cells transfected with siCTL or siAP-1γ1 (Fig. 3a). In WB, we used the 1E6 mAb that recognizes all the ORF2 isoforms. We also controlled the expression levels of tubulin and the stress-overexpressed GRP78 marker (Fig. 3a). In IP, we used P1H1 and P3H2 antibodies to differentially immunoprecipitate ORF2 forms in cell supernatants. Indeed, we have previously shown that the P1H1 mAb specifically immunoprecipitates the particle-associated ORF2i protein whereas the P3H2 mAb preferentially immunoprecipitates glycosylated ORF2g/c forms from heat-denatured HEV-cell culture supernatant [9]. Impact of the AP-1 silencing on viral production was also analyzed by quantification of viral RNAs (Fig. 3b) and infectious titers (Fig. 3c) and compared to cells treated with sofosbuvir (Sof), a well-characterized inhibitor of in vitro HEV replication and production [41].
The silencing of AP-1γ1 adaptin did not induce any cellular stress response (i.e. no impact on intracellular GRP78 detection levels in WB) and did not affect the intracellular expression level/pattern of ORF2 protein (Fig. 3a, Cells). In contrast, we found that the AP-1γ1 silencing induced a significant reduction of ORF2i protein detection in supernatants after immunoprecipitation with the anti-ORF2i P1H1 antibody, without affecting secretion of glycosylated ORF2g/c forms (Fig. 3a, Supernatant). These results suggest that the AP-1γ1 silencing likely reduces the secretion of viral particles. Consistently, the levels of extracellular viral RNA (Fig. 3b) and extracellular infectious viral particles (Fig. 3c) were reduced. Of note, intracellular HEV RNA levels were reduced by 40% upon AP-1γ1 silencing (Fig. 3b), indicating that the adaptor complex might be important for a viral or cellular factor involved in HEV replication. Indeed, the silencing of AP-1γ1 or AP-1γ2 adaptins in PLC3 cells stably replicating a p6 subgenomic replicon led to a 38% and 36% reduction in HEV RNA levels, respectively (Fig. S4), confirming that AP-1 complex plays a role in HEV replication. Strikingly, intracellular infectious titers were reduced by 80% upon AP-1γ1 silencing (Fig. 3c). These results indicate that the inhibition levels of intracellular viral RNA and infectious titers cannot be correlated and suggest that, in addition to an effect on HEV replication, AP-1γ1 silencing likely affects another step of the HEV life cycle, namely the assembly of infectious viral particles.
Taken together, these results highlight the importance of the AP-1 complex in HEV replication and addressing the ORF2i protein to viral factories for producing infectious viral particles. The AP-1 complex is probably a central player in the HEV lifecycle.
Pharmacological inhibition of the AP-1 complex prevents the localization of ORF2i protein in viral factories and inhibits viral infectivity
Since in AP-1 silencing experiments, we observed an effect on ORF2 subcellular localization, but also on HEV replication, we then wanted to use another approach to define the importance of AP-1 in the HEV lifecycle. We therefore used the pharmacological inhibitor A5 that specifically inhibits the AP-1 complex [25, 42, 43]. This compound is a cell-permeable piperazinyl phenylethanone derivate that specifically inhibits AP-1 complex by acting at a step after adaptor membrane recruitment, thereby significantly impairing AP-1 subcellular shuttling [42] (Fig. S5a). Therefore, we next sought to analyze the effect of this AP-1 traffic inhibitor, at non-cytotoxic dose of 150µM (Fig. S5b), on the subcellular localization of the ORF2i protein and the HEV lifecycle.
We first carried out an extensive immunofluorescence analysis of the impact of A5 treatment on HEV ORF2i protein subcellular localization and colocalization with viral/cellular markers, at 3 days post-treatment (Fig. 4), as described above.
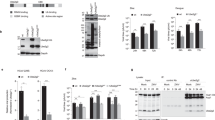
AP-1 complex pharmacological inhibition by A5 impairs ORF2i subcellular localization and colocalization with various viral/host cell markers. (a-g) At 6 d.p.e., PLC3/HEV cells were treated with H2O or A5 (150µM) and fixed after 3 days of treatment. Then, cells were permeabilized with cold methanol and 0.5% Triton X-100 and double-stained with the indicated antibodies. For each double staining, MOC of the ORF2i labelling in the cellular marker/ORF3 labelling was determined using the whole cell as ROI. Each data dot in the bar chart represents a cell field, and the total number of ORF2-positive cells used to calculate the MOC is indicated. Displayed results come from biological replicates. Mann-Whitney test, ****p < 0.0001
As shown in Fig. 4a, the ORF2i protein significantly colocalized with AP-1 complex in H2O-treated cells (MOC = 0.86) in perinuclear regions whereas this colocalization was significantly reduced in A5-treated cells (A5–150µM, MOC = 0.45). Interestingly, in A5-treated cells, ORF2i protein displayed a more diffuse subcellular distribution as compared to H2O-treated cells (Fig. 4a), similarly to what was observed in siAP-1γ1-transfected cells (Fig. 2a). As observed for the siAP-1γ1 transfection (Fig. S3a), we found that treatment with A5 compound had no impact on the subcellular localization of the AP-2 adaptor complex (Fig. S3b), confirming that this compound is specific to AP-1 complex. These results confirm the importance of the AP-1 complex in the subcellular addressing of ORF2i protein.
In line with silencing experiments, the M6PR marker also displayed a more diffuse cytosolic pattern and a reduced co-distribution with ORF2i (MOC = 0.49 in H2O-cells vs. 0.22 in A5-cells; Fig. 4b) upon treatment with A5, without any impact on TGN integrity (Fig. 4c-d), indicating that the A5 treatment alters AP-1 complex activity.
Clathrin also showed a more diffuse signal in the cytosol and reduced colocalization with ORF2i in A5-treated cells (MOC = 0.88 in H2O-cells vs. 0.50 in A5-cells; Fig. 4e). This indicates that AP-1 complex activity as well as clathrin recruitment are important for the subcellular addressing of ORF2i protein. Regarding ORF2i/ORF3 co-staining (Fig. 4f), ORF3 staining was less focalized in treated cells, with a significantly reduced MOC value in A5-treated cells (MOC = 0.22), as compared to siCTL cells (MOC = 0.80). This indicates that the inhibition of AP-1 complex alters HEV ORF2i/ORF3 proteins co-distribution. For ORF2i/Rab11 co-staining (Fig. 4g), a strong colocalization was observed in perinuclear nugget-like structures in the control condition (H2O, MOC = 0.82) whereas upon treatment with A5, the colocalization was significantly reduced (MOC = 0.42), indicating that pharmacological inhibition of AP-1 complex trafficking prevents the ORF2i protein from being targeted to viral factories. Altogether, these results confirm that AP-1 complex is an important host determinant involved in ORF2i protein addressing to viral factories.
Next, we sought to assess the impact of altered ORF2i protein localization in viral factories upon A5 treatment on the viral lifecycle. For that purpose, we proceeded as in Fig. 3 with PLC3/HEV cells treated with H2O or A5 (Fig. 5). As observed for the siRNA transfected cells, we found that the A5 treatment did not affect intracellular expression levels/patterns of ORF2 and ORF3 proteins (Fig. 5a, Cells). In addition, A5 treatment did not affect intracellular expression level/pattern of AP-1 complex and did not induce any cellular stress response (i.e. no impact on intracellular GRP78 detection levels in WB). Interestingly, we found that A5 treatment induced a significant reduction of ORF2i protein detection in supernatants after immunoprecipitation with the anti-ORF2i P1H1 antibody, without affecting secretion of glycosylated ORF2g/c forms (Fig. 5a, Supernatant). These results indicate that A5 treatment likely reduces the secretion of viral particles. Consistently, the levels of extracellular viral RNA (Fig. 5b) and extracellular infectious viral particles (Fig. 5c) were reduced. In contrast to siAP-1γ1-transfected cells (Fig. 3b), intracellular viral RNA levels were significantly increased upon A5 treatment (Fig. 5b). Since we observed that the treatment of PLC3 cells stably replicating a p6 subgenomic replicon with A5 compound only slightly reduced HEV RNA levels (Fig. S4), we hypothesized that the inhibition of secretion and/or assembly of viral particles by A5 leads to an intracellular accumulation of HEV RNA (Fig. 5b). Consistently, levels of intracellular infectious particles were significantly reduced upon A5 treatment (≈ 70%; Fig. 5c), indicating that A5 inhibits HEV assembly and consequently viral particle secretion. Taken together, these results indicate that inhibition of AP-1 complex activity impairs assembly and therefore secretion of infectious HEV particles.
AP-1 complex pharmacological inhibition by A5 alters viral RNA secretion and particle production. (a) Supernatants and cell lysates of PLC3/HEV/H2O, PLC3/HEV/A5-150µM, PLC3/H2O cells were generated after 3 days of treatment. In supernatants, ORF2i and ORF2g/c proteins were immunoprecipitated using anti-ORF2i P1H1 or anti-ORF2i/g/c P3H2 antibodies, respectively. An irrelevant mouse IgG antibody was used as an isotype control (Iso). ORF2 proteins were detected by WB using the 1E6 antibody. In cell lysates, ORF2i protein was detected by WB using 1E6 antibody. GRP78 and Tubulin proteins were detected using a rat anti-GRP78 antibody and a mouse anti-β-Tubulin antibody, respectively. (b) HEV RNA quantification in PLC3/HEV/H2O, PLC3/HEV/A5-150µM, PLC3/HEV/DMSO, PLC3/HEV/Sofosbuvir-20µM or PLC3 cells after 3 days of treatment. Extracellular and intracellular viral RNAs were quantified by RT-qPCR. Titers were adjusted to 100% for H2O/DMSO-treated cells. PLC3/HEV/Sofosbuvir-20µM cells were used as a positive control for replication inhibition. Values are from two independent experiments. Mann-Whitney test, **p < 0.01. (c) Infectious titer determination in PLC3/HEV/H2O, PLC3/HEV/A5-150µM, PLC3/HEV/DMSO, PLC3/HEV/Sofosbuvir-20µM or PLC3 cells after 3 days of treatment. Extracellular and intracellular viral particles were used to infect naïve Huh-7.5 cells for 3 days. Cells were next processed for indirect immunofluorescence. ORF2-positive cells were counted and each positive cell focus was considered as one FFU. Titers were adjusted to 100% for H2O/DMSO-treated cells. PLC3/HEV/Sofosbuvir-20µM cells were used as a positive control for infectious titers inhibition. Values are from three independent experiments. Mann-Whitney test, **p < 0.01, ****p < 0.0001
Taken together, these results confirm the importance of the AP-1 complex in addressing the ORF2i protein to viral factories and thus assembly and secretion of viral particles. The AP-1 complex is therefore a key player in the HEV lifecycle.
AP-1 complex-dependent shuttling is pivotal for the HEV lifecycle in PHHs
To further validate our findings in a cell culture system of hepatocytes closer to in vivo settings, we analyzed the impact of A5 inhibitor in HEV-infected primary human hepatocytes (PHHs) (Fig. 6). Briefly, one day after seeding (D0), PHHs were infected with HEV particles and treated with the compound A5, ribavirin (RBV, an FDA-approved HEV replication inhibitor), or their respective diluent (H2O or DMSO). At 3 days post-infection/treatment, supernatants were recovered (Fig. 6a). We assessed the impact of treatments on ORF2 protein detection in PHH supernatant by WB (Fig. 6b) and on viral production by quantification of infectious titers (Fig. 6c).
AP-1 complex is involved in particle production in primary human hepatocytes. (a) Experimental procedure. One day after seeding (D0), primary human hepatocytes (PHHs) were infected with intracellular HEV particles and treated with the appropriate drug/diluent. At D + 1 and D + 2, fresh medium containing drug/diluent was added to the wells. At D + 3, supernatants were recovered and processed for WB and extracellular infectious titers determination. D = Day. (b) Supernatant of HEV-infected PHHs or Mock PHHs, treated or not with diluent (H2O or DMSO), A5-150µM (A5) or Ribavirin-25µM (RBV) were subjected to western blotting to detect the ORF2g protein using the 1E6 Ab. The asterisk indicates a non-specific band detected in HEV-infected cells. (c) Supernatants were used to infect naïve Huh-7.5 cells for 3 days to determine extracellular infectious titers. Cells were next processed for indirect immunofluorescence. ORF2-positive cells were counted and each positive cell focus was considered as one FFU. Results were expressed in relative infectious titers. PHHs-Ribavirin-25µM (RBV) cells were used as a positive control for infectious titers inhibition. Values are from two independent experiments. Mann-Whitney test, **p < 0.01
The low amount of virus particles produced in the supernatants of infected PHH prevents the detection or immunoprecipitation of the ORF2i form, as carried out in Figs. 3 and 5. However, the ORF2g form, which is the predominant ORF2 form in cell culture supernatants [7, 8], can be easily detected by WB. This form follows the general secretion pathway, which is different from that of ORF2i protein [13]. As shown in Fig. 6b, A5 treatment did not affect the detection level/pattern of the ORF2g protein. In contrast, no ORF2g-associated signal was detected in the RBV-treated condition, due to RBV-induced replication inhibition. These results indicate that the A5 compound has not impact on the secretion of the ORF2g protein in PHH supernatant and, more globally, on the protein secretion of PHHs.
Next, we analyzed the impact of A5 treatment on viral production by determining the extracellular infectious titers (Fig. 6c). After treatment with A5, infectious titers were significantly reduced (≈ 40%) as compared to control conditions, to levels comparable to the RBV-treated condition (≈ 40%). This indicates that A5 treatment inhibits the production of viral particles in HEV-infected PHHs, as observed in hepatoma cell lines.
Taken together, these results indicate that AP-1-dependent shuttling is pivotal for the HEV particle secretion in this ex vivo system, without affecting the secretion of ORF2g protein. These results also confirm our conclusions drawn in hepatoma cell lines underlining the pivotal role of AP-1 complex in the HEV lifecycle.
HEV ORF2i protein interacts with the AP-1 adaptor complex
Next, we studied the interaction between the HEV ORF2i protein and the AP-1 complex (Fig. 7). First, we performed co-immunoprecipitation (co-IP) experiments in PLC3/HEV and PLC3 cells, at 6 d.p.e. (Fig. 7a). AP-1 complex was detected at similar intensities in PLC3/HEV and PLC3 cell lysates whereas ORF2i protein was only detected in PLC3/HEV cell lysate (“Inputs”, Fig. 7a). Following immunoprecipitation of AP-1 complex (IP AP-1), a faint band corresponding to the ORF2i protein was detected in PLC3/HEV cells (arrowhead) and not in PLC3 mock cells or in the isotype control condition (Iso), indicating that the ORF2i protein was specifically co-immunoprecipitated with AP-1 (Fig. 7a). Therefore, the AP-1 complex and the HEV ORF2i protein likely interact specifically in PLC3/HEV cells. Immunoprecipitation with anti-ORF2i P1H1 mAb was performed but failed to detect the AP-1 complex, probably because the P1H1 epitope is hidden by the interaction (data not shown).
HEV ORF2i protein interacts with AP-1 complex in hepatoma cell lines. (a) PLC3/HEV and PLC3 cell lysates were immunoprecipitated using a polyclonal anti-AP-1antibody (IP AP-1). An irrelevant rabbit IgG antibody was used as an isotype control (Iso). Inputs and immunoprecipitated AP-1 and ORF2 proteins were next detected by WB. (b) PLC3/HEV/H2O, PLC3/HEV/A5-150µM and PLC3/H2O cells were processed for proximity ligation assay using anti-ORF2i P1H1 and anti-AP-1γ1 antibodies after 3 days of treatment. Stacks of images corresponding to the total volume of the cells were acquired, and maximum intensity projections of the stacks were generated. For each condition, 12 fields of cells were analyzed (total cell number > 150 cells). Single stack of representative fields and quantification of spot/cell (bottom right) are shown. Kruskal-Wallis test, **p < 0.01, ****p < 0.0001. Molecular modeling of the interaction between ORF2i domain S and AP-1 complex subunit σ1. (c) Experimental overall structure of a truncated ORF2 form (aa 112–606) in the context of an icosahedral virus-like particle. The ordered domains S, M and P are colored in green, magenta and gray, respectively. The side chains of leucine 163 and leucine 164 are displayed as cyan spheres in (c) and (d). (d) AlphaFold2 modeling of a putative complex between the S domain (aa 131–312) of ORF2 (ORF2-S, green) and the subunit σ1A of AP-1 complex (AP-1 σ1, orange). (e) Predicted alignment error plot for the ORF2-S / σ1A complex. In this plot, residues are numbered sequentially (x-axis) so that residues 1-182 are ORF2-S and σ1 starts at 183, as indicated by black lines. The positioning error scale per predicted pair (in Å) is shown on the right. Blue dots indicate confident relative positioning of the residue pair considered. The overall blue color ORF2-S vs. σ1 (quadrants off the diagonal) indicates a confidently predicted structure of the ORF2-S / σ1 complex
In order to confirm our co-IP results, we next carried out proximity ligation assay (PLA) (Fig. 7b). PLA is a useful method to detect and visualize protein-protein interactions and post-translational modifications with high specificity and sensitivity. We used the Duolink PLA technology (Sigma) that uses a pair of primary antibodies and secondary antibodies labeled with oligonucleotides which when in close proximity (within 40 nm) undertake rolling circle amplification to generate specific fluorescent spot signals after the addition of labeled probes that can be visualized by microscopy [44]. To study ORF2i/AP-1 interaction, we used mouse anti-ORF2i P1H1 and rabbit anti-AP-1γ1 antibodies. In PLC3/HEV cells (HEV/H2O), a mean of 2.5 spots/cell was observed whereas less than 0.2 spot/cell were observed in mock electroporated cells (PLC3/H2O) (Fig. 7b), indicating that ORF2i protein and AP-1 complex are in close proximity and likely interact together. As adaptor proteins interact with their cargo proteins in a transient and low-affinity manner [45], the low number of spots per cell in PLC3/HEV cells may be explained by the transient interaction of the cargo protein/AP-1 complex in living cells, which would be in agreement with the low intensity of the ORF2i signal co-immunoprecipitated with AP-1 (Fig. 7a). Interestingly, when we treated the cells with the A5 compound (HEV/A5), we observed a significant reduction of spot detection (< 1 spot/cell) as compared to control cells (HEV/H2O) (Fig. 7b), confirming the specificity of ORF2i/AP-1 interaction.
Next, we used the breakthrough protein modeling program AlphaFold2 to assess how the ORF2i protein might interact with the AP-1 complex. We made systematic attempts with the human AP1 subunits β1, γ1, µ1A, µ1B, σ1A/σ1, σ1B/σ2 and σ1C/σ3. We did not obtain a reliable complex model, but we did find hints in the predicted alignment error (PAE) plots that there might be a propensity for interaction between AP-1 σ subunits and the shell (S) domain of ORF2 (ORF2-S). As such hints proved to be good indicators in our previous work on HEV ORF1 [46], we generated models with on the one hand ORF2-S (Fig. 7c) and on the other hand each of β1, γ1, both µ and all three σ subunits. In the case of the σ1A subunit, we obtained a definite hit (Fig. 7d-e), for which the model displays very low predicted alignment error statistics, even in the off-diagonal quadrant (ORF2i-S vs. AP-1σ1 and AP-1σ1 vs. ORF2i-S, Fig. 7e), a very reliable indicator of correct placement of two subunits in a protein-protein complex [47]. These results indicate that ORF2i likely recruits AP-1 via an interaction between its S domain and the σ1A/σ1 subunit. It is noteworthy that the interaction as modeled can only occur in a conformation of ORF2i distinct from that observed in self-assembled forms of ORF2, in which a conserved dileucine motif on the S domain (Fig. S6) that is involved in contact with σ1A/σ1 (Fig. 7d) is masked by the M domain (Fig. 7c). This again points to a transient interaction between ORF2i and AP-1.
Discussion
We and others demonstrated that during its lifecycle, HEV produces three different forms of the ORF2 capsid protein [7, 8]. The non-glycosylated ORF2i form is the structural component of viral particles. The ORF2g/c forms are massively secreted proteins that are N-glycosylated, O-glycosylated, sialylated but are not associated to virions. The mechanism underlying the differential production of these isoforms is not fully understood but we have recently shown that an arginine-rich motif (ARM) located at the N-terminus of the ORF2 protein, is responsible for regulating the ORF2 protein topology toward secretion pathway membranes to produce either ORF2i or ORF2g/c forms. We showed that the ORF2i protein is likely anchored into the membranes through its N-terminus and faces the cytosol [13]. More recently, using electron microscopy approaches and the P1H1 anti-ORF2i specific antibody, we have shown that HEV induces tubular and vesicular structures in the vicinity of the nucleus that are enriched in viral proteins (i.e., ORF1, ORF2i, ORF3 proteins), viral RNA and ERC resident proteins (i.e., Rab11, CD71) [9, 48]. In confocal microscopy, these structures form nugget-like structures in perinuclear areas. Importantly, silencing of Rab11 significantly impaired viral particle production in a replication-independent manner and altered the subcellular localization of ORF2i protein [9]. These data suggest that, in addition to multivesicular bodies which have been shown to be instrumental in the late steps of the HEV lifecycle [49, 50], the ERC likely serves as a viral factory and also constitutes a pathway for viral particle egress. One of the questions underlying these results is the mechanism responsible for driving viral elements (proteins and RNA) to ERC/viral factories, and more specifically the ORF2i protein, a key player in the HEV lifecycle.
In the present study, we have demonstrated the importance of the AP-1 adaptor complex in addressing the ORF2i protein to ERC/viral factories. Both proteins colocalized and interacted together in hepatoma cell lines. Inhibition of AP-1 complex by silencing or by a pharmacological inhibitor impaired this interaction and significantly reduced viral infectivity, underlining the importance of the interplay between AP-1 and ORF2i for producing viral particles, in hepatoma cell lines and primary human hepatocytes (Fig. 8). The interaction between AP-1 complex and viral proteins has been shown to be important for infectivity of several viruses belonging to different families. For example, AP-1 complex interacts with HIV Nef [51], African swine fever virus CD2v [52] and Varicella-Zoster ORF9p [23].
Model of ORF2i protein addressing to viral factories by AP-1 complex. Newly produced ORF2i proteins are anchored in secretion pathway membranes through their Nter and oriented toward the cytosol. ORF2i protein then interacts with AP-1 complex in TGN to generate ORF2i protein/AP-1 complex-positive transport vesicles, in a clathrin dependent-manner. The transport vesicles transit to the endosomal recycling compartment (ERC) that constitutes viral factories where ORF2i protein colocalizes with the ERC resident marker Rab11. The enrichment of ORF1, ORF2i, ORF3 proteins and viral RNA in viral factories leads to viral assembly and production/secretion of infectious HEV particles. The inhibition of AP-1 complex activity either by A5 molecule or by silencing the AP-1γ1 adaptin (siAP-1γ1) prevents the ORF2i protein/AP-1 complex interaction, resulting in the absence of AP-1-positive vesicles containing ORF2i protein. In this context, ORF2i protein is no longer addressed to ERC/viral factories and does not colocalize with Rab11. Such process likely interferes with viral assembly, resulting in a significant reduction of infectious HEV particles production/secretion and ORF2i protein detection in culture supernatant
The interaction of AP-1 complex with cargoes relies on particular consensus sequences located in their cytoplasmic domain: the tyrosine-based motif YXXØ or the dileucine-based motif [DE]XXXL[LI] (X is any amino acid and Ø represents hydrophobic amino acid) [15]. Tyrosine-based motifs interact with AP-1 complex through the binding to the µ1 subunits whereas dileucine-based motifs bind to the combination of γ-σ1 subunits [15]. Here, molecular modeling analyses revealed that the OR2i protein likely interacts with AP-1 through an interaction between the ORF2 S domain and the subunit σ1A of AP-1. We therefore analyzed the primary ORF2 protein sequence and identified a fully conserved dileucine-like motif located in the ORF2 S domain (Fig. 7c-d and Fig. S6). This motif is conserved among strains isolated from human (gt1-4), wild boar (wb), pig (sw), rabbit (rab), and rat (Fig. S6). We performed site-directed mutagenesis of this dileucine-like motif in the HEV-p6 strain and characterized the generated mutant (DL mutant, Fig. S7). In line with what we observed when inhibiting the AP-1 complex, subcellular distribution of the DL mutant and its co-localization with AP-1, ORF3 and Rab11 were affected (Fig. S7b), as compared with the wildtype ORF2i protein. Moreover, infectivity and secretion of extracellular RNA of the DL mutant were reduced (Fig. S7d). However, further characterization of the latter by IP using a conformation-specific anti-ORF2 antibody (4B2, Fig. S7c) revealed that mutation of the dileucine motif likely results in some degree of misfolding of the ORF2 protein, making the results difficult to interpret. Further studies are therefore needed.
It has been shown that the TGN resident protein TGOLN2/TGN46 can be found on the surface of enveloped HEV particles, suggesting that they bud from intracellular vesicles likely originating from the TGN compartment [53, 54]. As AP-1 complex binds cargos anchored in TGN membranes, our study is likely in line with these observations. A study has also shown that ORF3 protein is palmitoylated on 8 N-terminal cysteine residues, and is exposed on the cytosolic side of intracellular membranes [14]. The ORF3 protein has also been found localized in ERC [9, 55]. As ORF2i and ORF3 proteins display the same topology towards intracellular membranes and are in close relationship, we cannot exclude that the ORF3 protein is also addressed to ERC/viral factories in an AP-1 complex-dependent manner, or that ORF2i-ORF3 protein complex is transported by the AP-1 complex. The fact that the subcellular localization of ORF3 protein is altered after inhibition of AP-1 complex supports this hypothesis. In addition to post-translational modifications of cargos, specific cytoplasmic proteins recruited as accessory proteins may be involved in AP-1 binding to facilitate cargo recruitment, such as the Phosphofurin Acidic Cluster Sorting Protein (PACS) family, including PACS-1 [56, 57]. We can speculate that ORF2i protein may be involved, for example, in ensuring the proper localization of the ORF3 protein, as PACS-1 does. In this context, inhibition of the ORF2i protein/AP-1 complex interaction would alter ORF3 protein subcellular localization and viral assembly/secretion: these results were observed in our study and thus support this hypothesis. Another explanation would have been that inhibition of AP-1 complex (by A5 or silencing of AP-1γ1) directly affects the ORF3 protein and not the ORF2i protein. However, by using cells expressing only the ORF2 protein (ORF2-p6 or ORF2-Sar55), we showed that the ORF2i protein colocalizes with AP-1 complex in the absence of ORF3 protein. In addition, the absence in the ORF3 protein sequence of dileucine-based motif [DE]XXXL[LI] or tyrosine-based motif YXXØ, which are important consensus sequences in cargoes for their interaction with AP-1 complex, makes it unlikely that ORF3 interacts with AP-1 complex. Moreover, previous characterization of HEVΔORF3 virus, expressing an ORF3-null mutant of HEV-p6, showed that virion secretion was affected while viral assembly was not, indicating that the ORF3 protein is necessary for viral particle secretion but not for viral assembly. Specifically, extracellular infectious titers were significantly reduced compared to wild-type virus [58]. In the present study, we observed an ≈ 70% reduction in extracellular and intracellular infectious titers in cells treated with A5 or transfected with siRNA targeting AP-1γ1, indicating that viral assembly was altered. Therefore, our observations cannot be attributed to an effect on the ORF3 protein, but are specific to the ORF2i protein.
Unexpectedly, the interaction between the ORF2i protein and AP-1 complex may be of greater importance. As AP-1 complex is involved in many cellular protein transport processes, this interaction could disrupt the delivery and/or subcellular addressing of cellular proteins. This could play a role in promoting viral infection, interfering with the recruitment of restriction factors or evading host immune responses. For example, in the context of HIV infection, the interaction of Nef viral protein with AP-1 complex interferes with the subcellular addressing of cellular proteins, notably by decreasing the amount of class I MHC molecules or CD4 glycoprotein detectable on the cell surface [59, 60]. This mechanism is likely involved in the immune escape of HIV-infected cells, mainly by inhibiting the presentation of viral antigens. This aspect would deserve our attention in order to better understand the effect of the ORF2i/AP-1 complex interaction in the context of HEV infection at the cellular level. Further characterization of this interaction could be helpful to identify new host factors involved in promoting viral infection as well as new restriction factors or to understand immune evasion mechanisms settled up by HEV, thus revealing potential new drug targets for combating HEV infection.
Our results showed that AP-1 complex silencing reduces HEV replication, indicating that homeostasis of endosomal pathway is likely to be important for HEV replication. In line with this, we previously found that the guanine-exchange factor (GEF) GBF1 is necessary to HEV replication but has an indirect role by activating effectors involved in HEV replication [61]. GEFs regulate the activation of Arf proteins that are small G-proteins regulating the recruitment of adaptor proteins to the membranes, in particular Arf1 drives the membrane recruitment of AP-1 complex [38]. Interestingly, a very recent study confirmed GBF1 as a key factor in HEV replication and highlighted a role of Rab5A and the importance of endosomal membrane homeostasis in HEV replication [62]. Of note, the less marked effect of compound A5 on HEV replication is likely due to the fact that this compound only blocks AP-1-dependent trafficking between the TGN and endosomes without altering other trafficking pathways [42].
In conclusion, we have identified for the first time a host determinant critical for the HEV lifecycle that is involved in HEV ORF2i protein localization to ERC/viral factories. In addition to expanding the scarce knowledge of host factors involved in the HEV lifecycle, we have demonstrated that AP-1-dependent shuttling is important for HEV particle production in human hepatoma cells in vitro or ex vivo in primary human hepatocytes.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AP:
-
Adaptor protein complex
- AP:
-
1-Adaptor protein complex 1
- ARM:
-
Arginine-rich motif
- CTL:
-
Control
- d.p.e:
-
Days post-electroporation
- d.p.i:
-
Days post-infection
- EM:
-
Electron microscopy
- ERC:
-
Endocytic recycling compartment
- GRP78:
-
78 kDa glucose-regulated protein
- Gt:
-
Genotype
- HD:
-
Heat denatured
- HEV:
-
Hepatitis E virus
- HIV:
-
Human immunodeficiency virus
- h.p.t.:
-
Hours post-transfection
- IF:
-
Immunofluorescence
- IP:
-
Immunoprecipitation
- Iso:
-
Isotype control
- M6PR:
-
Mannose-6-phosphate receptor
- MOC:
-
Mander’s overlapping coefficient
- NT:
-
Non-transfected
- ORF:
-
Open reading frame
- ORF2c:
-
Cleaved ORF2
- ORF2g:
-
Glycosylated ORF2
- ORF2i:
-
Infectious ORF2
- PAE:
-
Predicted alignment error
- PHHs:
-
Primary human hepatocytes
- PLA:
-
Proximity ligation assay
- RBV:
-
Ribavirin
- ROI:
-
Region of interest
- SARS:
-
CoV-2-Severe Acute Respiratory Syndrome-Coronavirus-2
- siRNA:
-
Small interfering RNA
- SN:
-
Supernatant
- Sof:
-
Sofosbuvir
- STX6:
-
Syntaxin-6
- TGN:
-
Trans-Golgi network
- TGN46:
-
Trans-Golgi network 46 kDa protein
- WB:
-
Western-blotting
References
Purdy MA, Drexler JF, Meng X-J et al (2022) ICTV Virus Taxonomy Profile: Hepeviridae 2022. J Gen Virol 103. https://doi.org/10.1099/jgv.0.001778
Kamar N, Izopet J, Pavio N et al (2017) Hepatitis E virus infection. Nat Reviews Disease Primers 3:17086. https://doi.org/10.1111/trf.13355
Ahmed R, Nasheri N (2023) Animal reservoirs for hepatitis E virus within the Paslahepevirus Genus. Vet Microbiol 278:109618. https://doi.org/10.1016/j.vetmic.2022.109618
Lhomme S, Marion O, Abravanel F et al (2020) Clinical manifestations, Pathogenesis and treatment of Hepatitis E Virus infections. J Clin Med 9:331. https://doi.org/10.3390/jcm9020331
Pischke S, Hartl J, Pas SD et al (2017) Hepatitis E virus: infection beyond the liver? J Hepatol 66:1082–1095. https://doi.org/10.1016/j.jhep.2016.11.016
Nimgaonkar I, Ding Q, Schwartz RE, Ploss A (2018) Hepatitis E virus: advances and challenges. Nat Reviews Gastroenterol Hepatol 15:96–110. https://doi.org/10.1038/nrgastro.2017.150
Montpellier C, Wychowski C, Sayed IM et al (2018) Hepatitis E Virus Lifecycle and Identification of 3 forms of the ORF2 capsid protein. Gastroenterology 154:211–223e8. https://doi.org/10.1053/j.gastro.2017.09.020
Yin X, Ying D, Lhomme S et al (2018) Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc Natl Acad Sci USA 3:201721345–201721346. https://doi.org/10.1073/pnas.1721345115
Bentaleb C, Hervouet K, Montpellier C et al (2022) The endocytic recycling compartment serves as a viral factory for hepatitis E virus. Cell Mol Life Sci 79:615. https://doi.org/10.1007/s00018-022-04646-y
Ankavay M, Montpellier C, Sayed IM et al (2019) New insights into the ORF2 capsid protein, a key player of the hepatitis E virus lifecycle. Sci Rep 9:6243. https://doi.org/10.1038/s41598-019-42737-2
Nilsson J, Persson B, von Heijne G (2005) Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins 60:606–616. https://doi.org/10.1002/prot.20583
von Heijne G (1989) Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341. https://doi.org/10.1038/341456a0
Hervouet K, Ferrié M, Ankavay M et al (2022) An arginine-rich motif in the ORF2 capsid protein regulates the hepatitis E virus lifecycle and interactions with the host cell. Plos Pathog 18:e1010798. https://doi.org/10.1371/journal.ppat.1010798
Gouttenoire J, Pollán A, Abrami L et al (2018) Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PLoS Pathog 14:e1007471. https://doi.org/10.1371/journal.ppat.1007471
Park SY, Guo X (2014) Adaptor protein complexes and intracellular transport. Bioscience Rep 34:e00123. https://doi.org/10.1042/bsr20140069
Robinson MS, Bonifacino JS (2001) Adaptor-related proteins. Curr Opin Cell Biol 13:444–453. https://doi.org/10.1016/s0955-0674(00)00235-0
Hirst J, Barlow LD, Francisco GC et al (2011) The Fifth adaptor protein complex. PLoS Biol 9:e1001170. https://doi.org/10.1371/journal.pbio.1001170
Shin J, Nile A, Oh J-W (2021) Role of adaptin protein complexes in intracellular trafficking and their impact on diseases. Bioengineered 12:8259–8278. https://doi.org/10.1080/21655979.2021.1982846
Duncan MC (2022) New directions for the clathrin adaptor AP-1 in cell biology and human disease. Curr Opin Cell Biol 76:102079. https://doi.org/10.1016/j.ceb.2022.102079
Wang YJ, Wang J, Sun HQ et al (2003) Phosphatidylinositol 4 phosphate regulates targeting of Clathrin adaptor AP-1 complexes to the Golgi. Cell 114:299–310. https://doi.org/10.1016/s0092-8674(03)00603-2
Baust T, Czupalla C, Krause E et al (2006) Proteomic analysis of adaptor protein 1A coats selectively assembled on liposomes. Proc Natl Acad Sci 103:3159–3164. https://doi.org/10.1073/pnas.0511062103
Ren X, Farías GG, Canagarajah BJ et al (2013) Structural Basis for Recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 152:755–767. https://doi.org/10.1016/j.cell.2012.12.042
Lebrun M, Lambert J, Riva L et al (2018) Varicella-Zoster Virus ORF9p binding to Cellular adaptor protein complex 1 is important for viral infectivity. J Virol 92. https://doi.org/10.1128/jvi.00295-18
Rebendenne A, Roy P, Bonaventure B et al (2022) Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. Nat Genet 54:1090–1102. https://doi.org/10.1038/s41588-022-01110-2
Yasamut U, Tongmuang N, Yenchitsomanus P et al (2015) Adaptor protein 1A facilitates Dengue Virus Replication. PLoS ONE 10:e0130065. https://doi.org/10.1371/journal.pone.0130065
Lenggenhager D, Gouttenoire J, Malehmir M et al (2017) Visualization of hepatitis E virus RNA and proteins in the human liver. J Hepatol 67:471–479. https://doi.org/10.1016/j.jhep.2017.04.002
Jothikumar N, Cromeans TL, Robertson BH et al (2006) A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131:65–71. https://doi.org/10.1016/j.jviromet.2005.07.004
Mirdita M, Schütze K, Moriwaki Y et al (2022) ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. https://doi.org/10.1038/s41592-022-01488-1
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Mirdita M, Steinegger M, Söding J (2019) MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35:2856–2858. https://doi.org/10.1093/bioinformatics/bty1057
Steinegger M, Söding J (2017) MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35:1026–1028. https://doi.org/10.1038/nbt.3988
Madeira F, Pearce M, Tivey ARN et al (2022) Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50:W276–W279. https://doi.org/10.1093/nar/gkac240
Romero-Brey I, Merz A, Chiramel A et al (2012) Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. https://doi.org/10.1371/journal.ppat.1003056
Klumperman J, Hille A, Veenendaal T et al (1993) Differences in the endosomal distributions of the two mannose 6-phosphate receptors. J cell Biol 121:997–1010. https://doi.org/10.1083/jcb.121.5.997
Ghosh P, Dahms NM, Kornfeld S (2003) Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol 4:202–213. https://doi.org/10.1038/nrm1050
Doray B, Ghosh P, Griffith J et al (2002) Cooperation of GGAs and AP-1 in packaging MPRs at the Trans-Golgi Network. Science 297:1700–1703. https://doi.org/10.1126/science.1075327
Ghosh P, Kornfeld S (2004) The cytoplasmic tail of the cation-independent mannose 6-phosphate receptor contains four binding sites for AP-1. Arch Biochem Biophys 426:225–230. https://doi.org/10.1016/j.abb.2004.02.011
Tan JZA, Gleeson PA (2019) Cargo sorting at the Trans-Golgi Network for shunting into specific transport routes: role of Arf Small G Proteins and adaptor complexes. Cells 8:531. https://doi.org/10.3390/cells8060531
Luzio JP, Brake B, Banting G et al (1990) Identification, sequencing and expression of an integral membrane protein of the trans -golgi network (TGN38). Biochem J 270:97–102. https://doi.org/10.1042/bj2700097
Bock JB, Klumperman J, Davanger S, Scheller RH (1997) Syntaxin 6 functions in trans-golgi network vesicle trafficking. Mol Biol Cell 8:1261–1271. https://doi.org/10.1091/mbc.8.7.1261
Thi VLD, Debing Y, Wu X et al (2016) Sofosbuvir inhibits Hepatitis E Virus Replication in Vitro and results in an Additive Effect when combined with ribavirin. Gastroenterology 150:82–85e4. https://doi.org/10.1053/j.gastro.2015.09.011
Duncan MC, Ho DG, Huang J et al (2007) Composite synthetic lethal identification of membrane traffic inhibitors. Proc Natl Acad Sci 104:6235–6240. https://doi.org/10.1073/pnas.0607773104
Zallocchi M, Delimont D, Meehan DT, Cosgrove D (2012) Regulated vesicular trafficking of specific PCDH15 and VLGR1 variants in auditory hair cells. J Neurosci 32:13841–13859. https://doi.org/10.1523/jneurosci.1242-12.2012
Alam MS (2018) Proximity ligation assay (PLA). Curr Protoc Immunol 123:e58. https://doi.org/10.1002/cpim.58
Traub LM, Bonifacino JS (2013) Cargo Recognition in Clathrin-Mediated Endocytosis. Cold Spring Harb Perspect Biol 5:a016790. https://doi.org/10.1101/cshperspect.a016790
Fieulaine S, Tubiana T, Bressanelli S (2023) De novo modelling of HEV replication polyprotein: five-domain breakdown and involvement of flexibility in functional regulation. Virology 578:128–140. https://doi.org/10.1016/j.virol.2022.12.002
Schweke H, Pacesa M, Levin T et al (2024) An atlas of protein homo-oligomerization across domains of life. Cell 187:999–1010e15. https://doi.org/10.1016/j.cell.2024.01.022
Metzger K, Bentaleb C, Hervouet K et al (2022) Processing and subcellular localization of the Hepatitis E Virus Replicase: identification of candidate viral factories. Front Microbiol 13:828636. https://doi.org/10.3389/fmicb.2022.828636
Nagashima S, Jirintai S, Takahashi M et al (2014) Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol 95:2166–2175. https://doi.org/10.1099/vir.0.066910-0
Feng Z (2020) Quasi-enveloped Hepatitis virus assembly and release. Adv Virus Res 108:315–336. https://doi.org/10.1016/bs.aivir.2020.08.004
Bresnahan PA, Yonemoto W, Ferrell S et al (1998) A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol 8:1235–S1. https://doi.org/10.1016/s0960-9822(07)00517-9
Pérez-Núñez D, García-Urdiales E, Martínez-Bonet M et al (2015) CD2v interacts with adaptor protein AP-1 during African swine fever infection. PLoS ONE 10:e0123714. https://doi.org/10.1371/journal.pone.0123714
Nagashima S, Takahashi M, Jirintai S et al (2014) The membrane on the surface of hepatitis E virus particles is derived from the intracellular membrane and contains trans-golgi network protein 2. Arch Virol 159:979–991. https://doi.org/10.1007/s00705-013-1912-3
Das A, Rivera-Serrano EE, Yin X et al (2023) Cell entry and release of quasi-enveloped human hepatitis viruses. Nat Rev Microbiol 1–17. https://doi.org/10.1038/s41579-023-00889-z
Chandra V, Kar-Roy A, Kumari S et al (2008) The Hepatitis E Virus ORF3 protein modulates epidermal growth factor receptor trafficking, STAT3 translocation, and the Acute-Phase response. J Virol 82:7100–7110. https://doi.org/10.1128/jvi.00403-08
Wan L, Molloy SS, Thomas L et al (1998) PACS-1 defines a Novel Gene Family of Cytosolic sorting proteins required for trans-golgi network localization. Cell 94:205–216. https://doi.org/10.1016/s0092-8674(00)81420-8
Tu Y, Zhao L, Billadeau DD, Jia D (2020) Endosome-to-TGN trafficking: organelle-vesicle and organelle-organelle interactions. Front Cell Dev Biol 8:163. https://doi.org/10.3389/fcell.2020.00163
Emerson SU, Nguyen H, Torian U, Purcell RH (2006) ORF3 protein of Hepatitis E Virus is not required for replication, Virion Assembly, or infection of Hepatoma cells in Vitro. J Virol 80:10457–10464. https://doi.org/10.1128/jvi.00892-06
Iijima S, Lee Y-J, Ode H et al (2012) A noncanonical mu-1A-Binding motif in the N terminus of HIV-1 Nef determines its ability to Downregulate Major Histocompatibility Complex Class I in T lymphocytes. J Virol 86:3944–3951. https://doi.org/10.1128/jvi.06257-11
Tavares LA, de Carvalho JV, Costa CS et al (2020) Two functional variants of AP-1 complexes composed of either γ2 or γ1 subunits are independently required for Major Histocompatibility Complex Class I downregulation by HIV-1 Nef. J Virol 94. https://doi.org/10.1128/jvi.02039-19
Farhat R, Ankavay M, Lebsir N et al (2018) Identification of GBF1 as a cellular factor required for hepatitis E virus RNA replication. Cell Microbiol 20. https://doi.org/10.1111/cmi.12804
Oechslin N, Silva ND, Ankavay M et al (2023) A genome-wide CRISPR/Cas9 screen identifies a role for Rab5A and early endosomes in hepatitis E virus replication. Proc Natl Acad Sci 120:e2307423120. https://doi.org/10.1073/pnas.2307423120
Acknowledgements
We would like to thank Suzanne U. Emerson (NIH, United States), Jérôme Gouttenoire (University Hospital of Lausanne, Switzerland) and Ralph Bartenschlager (University of Heidelberg, Germany) for providing us with reagents. We also thank Audrey Tarricone and Sophana Ung for their technical assistance. We would also like to thank the imaging core facility of the BioImaging Center Lille-Nord de France for access to the infrastructure, and the integrative bioinformatics BIOI2 facility for making the ColabFold pipeline easily accessible at I2BC.
Funding
MF was supported by a fellowship from the Institut Pasteur de Lille, the Région Hauts-de-France and the ANRS-Maladies infectieuses émergentes (ANRS-MIE). TT was supported by a postdoctoral fellowship from ANRS-MIE. LM is currently supported by a Région Hauts-de-France/University of Lille fellowship. MA was supported by ANRS-MIE and is currently supported by the Fonds National Suisse (FNS). CB was supported by ANRS-MIE, INSERM-Transfert, Région Hauts-de-France and Institut Pasteur de Lille. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, experiments, data collection and analysis were performed by MF, VA, CM, PB, TT, LM, MA, CB, JD, SB, CMA, YR and LC. MF and LC wrote the first draft of the manuscript, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study does not include any human subjects.
Consent for publication
All authors read and approved the final manuscript.
Competing interests
Claire Montpellier, Jean Dubuisson and Laurence Cocquerel are coinventors of two patent applications on the use of antibodies having specificity for the ORF2i protein for HEV diagnostic purposes. Patents have been filed by Inserm Transfert. The authors have no other financial or non-financial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ferrié, M., Alexandre, V., Montpellier, C. et al. The AP-1 adaptor complex is essential for intracellular trafficking of the ORF2 capsid protein and assembly of Hepatitis E virus. Cell. Mol. Life Sci. 81, 335 (2024). https://doi.org/10.1007/s00018-024-05367-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05367-0