Abstract
P38γ (MAPK12) is predominantly expressed in triple negative breast cancer cells (TNBC) and induces stem cell (CSC) expansion resulting in decreased survival of the patients due to metastasis. Abundance of G-rich sequences at MAPK12 promoter implied the functional probability to reverse tumorigenesis, though the formation of G-Quadruplex (G4) structures at MAPK12 promoter is elusive. Here, we identified two evolutionary consensus adjacent G4 motifs upstream of the MAPK12 promoter, forming parallel G4 structures. They exist in an equilibria between G4 and duplex, regulated by the binding turnover of Sp1 and Nucleolin that bind to these G4 motifs and regulate MAPK12 transcriptional homeostasis. To underscore the gene-regulatory functions of G4 motifs, we employed CRISPR-Cas9 system to eliminate G4s from TNBC cells and synthesized a naphthalene diimide (NDI) derivative (TGS24) which shows high-affinity binding to MAPK12-G4 and inhibits MAPK12 transcription. Deletion of G4 motifs and NDI compound interfere with the recruitment of the transcription factors, inhibiting MAPK12 expression in cancer cells. The molecular basis of NDI-induced G4 transcriptional regulation was analysed by RNA-seq analyses, which revealed that MAPK12-G4 inhibits oncogenic RAS transformation and trans-activation of NANOG. MAPK12-G4 also reduces CD44High/CD24Low population in TNBC cells and downregulates internal stem cell markers, arresting the stemness properties of cancer cells.
Similar content being viewed by others
Introduction
The mitogen-activated protein kinase (MAPK) cascade is an essential signaling pathway, in which the input signals from various upstream stimuli converge to promote tumorigenesis by inducing tumor-suppressive and pro-oncogenic effects, thus leading to cancer cell survival, stemness, dissemination, metastasis, and therapeutic resistance. Although extensive research has been conducted to characterize and understand the functions of the MAPK superfamily of proteins, namely MAPK14 (p38α), MAPK11 (p38β), MAPK12 (p38γ), and MAPK13 (p38δ) [1, 2], the physiological and pathological functions of MAPK12 (p38γ) remain largely unaddressed. MAPK12 (aka ERK3; ERK6; ERK-6; SAPK3; PRKM12; SAPK-3; MAPK 12; P38GAMMA) was initially reported to have abundant expression in the skeletal muscles. However, the recent development of the global and tissue-specific MAPK12 (p38γ)-knockout mouse lines, acknowledged that MAPK12 expression is not as restricted as initially thought and exhibits pleiotropic cellular functions through cytokine production, cell cycle regulation, exocytosis, migration, invasion, differentiation, and neuron activity; having acute pathophysiological functions related to inflammation [3], cancer stemness [4,5,6], tumorigenesis, diabetes, neurodegeneration, cardiac hypertrophy, and glucose homeostasis. Aberrant expression of MAPK12 has recently been identified as the diagnostic and prognostic marker in triple-negative breast cancer (TNBC), glioma malignancy, colon cancer, and non-small cell lung cancer (NSCLC), associated clinically with lower overall patient survival [7].
Previous studies have shown that MAPK12 expression is highly induced by RAS activation, which positively regulates phosphorylation-independent oncogenic RAS transformation in various cancers [8,9,10,11,12,13]. Alternatively, MAPK12 downregulation arrests oncogenic RAS transformation, which leads to the diminution of metastatic and oncogenic properties of cancer cells [14,15,16,17]. In breast cancer, MAPK12-induced KRAS transformation integrates with estrogen receptor-mediated pathways to promote tumorigenesis and invasion [18, 19]. In colorectal cancer, MAPK12 induces KRAS-mediated invasion and metastasis, leading to the transactivation of MMP9 (Matrix metalloprotease 9). In pancreatic cancer, MAPK12 mediates the crosstalk between RAS signaling and aerobic glycolysis [9], suggesting MAPK12-induced RAS transformation as the crucial step to activate the signaling cascades leading to tumorigenesis, stemness, metastasis, and cell invasion in cancer. Lately, MAPK12 activation has been shown to promote cancer stemness properties by inhibiting microRNA-200b turnover through the ubiquitin-mediated proteasomal degradation of GATA3 [4]. In a more recent study, MAPK12 has been reported to regulate G0-to-G1 transition by inducing retinoblastoma phosphorylation that enables the cells to escape from the quiescent stage in response to the stress stimuli, playing a significant role in hepatocyte regeneration after partial hepatectomy [20]. Other investigations indicated the involvement of MAPK12 in myoblast differentiation [21] and in regulating cytoskeleton integrity [22]. This envisaged that MAPK12 is one of the key proteins to regulate the acquisition of stem cell-like traits in the cancer cells. Besides, MAPK12 activation stimulates the pro-inflammatory cytokines that play a central role in the development of inflammation-induced neoplastic processes, fostering proliferation and metastasis. With the revelation of these oncogenic functions, emerging studies are being made to produce specific MAPK12 inhibitors [23]. SB203580 and BIRB0796 are two well-established specific inhibitors for MAPK12 [24]. However, these compounds exhibit promiscuous binding to MAPK14 that expresses ubiquitously in all cell types, resulting in off-target effects. SU005 has recently been identified as a specific MAPK12 inhibitor, utilizing the structural differences between the active forms of MAPK14 and MAPK12 gatekeeper residues at the hinge region of the ATP-binding pocket [25, 26]. Although SU005 inhibits MAPK12 expression in the cultured cells, it binds to MAPK13, another isoform of MAPK superfamily, having off-target effects [27]. Recently, an anti-inflammatory drug, pirfenidone has been used as MAPK12-specific inhibitor, while its specificity for other MAPK isoforms remains to be tested. Therefore, the lack of MAPK12-specific inhibitors and its implications in tissue regeneration, cancer, and metabolic diseases, necessitate the development of novel therapeutic strategies targeting MAPK12 [28, 29].
G-Quadruplex (G4) nucleic acids have long been identified as attractive therapeutic targets in anticancer research [30]. These noncanonical DNA secondary structures are formed over the tandem repeats of guanine (G)-rich sequences having multiple π–π stacks of G-tetrads. These cohere through Hoogsteen hydrogen bonds involving N1, N7, O6, and N2 of G bases. The central carbonyl O6 offers G4 stability through metal cation (Na+ and K+) coordination [31]. G4s have received considerable attention in the past due to their widespread distribution in human genome, viz. telomeres, promoters, open reading frames, untranslated regions of the oncogenes, and translocation hotspots [32, 33]. Genome-wide analyses have further suggested that G4 structures are concentrated within the functional genomic regions, from highly divergent organisms, and play imperative roles in myriad gene-regulatory functions, such as transcription, replication, telomere maintenance, and translation regulation [34,35,36,37]. Although progress has been made in understanding G4 structures and their formation, their precise functional significance, and the impact of their conformational dynamics on transcriptional regulation are still areas of active research and investigation. Previous studies have revealed the importance of full conformational ensemble of G4 motifs for their wild-type functions [38], bringing into question the experimental strategies that utilize designed G4 sequences, tailored to predominantly populate single-ground conformations. Most of the G4-targeting compounds fail to conform to the polymorphic skeleton of wild-type G4s and suffer from promiscuous binding to other G4s, resulting in off-target effects. Earlier investigations reported that MAPK12 transcription is stringently regulated by the G-rich elements in its promoter that bind to Sp1 in a sequence-dependent manner, leading to the upregulation of MAPK12 transcription [39]. Since Sp1 has been widely studied as a potential G4 binding protein, the possibility that these G-rich domains at MAPK12 promoter fold into G4 structures is conspicuous. In a more recent study from Neidle’s group revealed the enrichment of putative G4 sequences (PQS) at MAPK12 promoter, having possible functions to reverse tumorigenesis in gemcitabine-resistant pancreatic cancer cells, suggesting the PQS in MAPK12 promoter as an attractive therapeutic target [40].
In this study, we identified two adjacently located G4 motifs at MAPK12 promoter that poise homeostatic regulation in MAPK12 transcription. They exhibit equilibria between duplex and G4 forms, regulated by the binding turnover of Nucleolin and Sp1 at tandem G4 elements. We harnessed CRISPR-Cas9-mediated deletion of these MAPK12-G4s to underscore their gene-regulatory functions in MAPK12-mediated oncogenesis. We also developed the chemical derivatives of naphthalene diimide (NDI) that induce and stabilize G4 formation at MAPK12 promoter and abrogate G4-mediated MAPK12 transcription in breast cancer cells. The molecular basis of NDI-induced G4 regulation was accessed by transcriptome-wide analyses using RNA-seq that revealed the inhibitory functions of tandem MAPK12-G4 motifs in oncogenic RAS transformation, which in turn thwart the transactivation of NANOG. G4 formation at MAPK12 promoter also reduces CD44High/CD24Low population in breast cancer cells and downregulates internal stem cell markers. The conformational dynamics between two neighboring G4s in MAPK12 promoter plays a role in diminishing the stem cell-like characteristics of breast cancer cells. Additionally, they link MAPK12's effects on RAS transformation to pathways that control stem cell-like behavior.
Materials and methods
Preparation of oligonucleotide sequences
Putative G4-forming sequences at MAPK12 promoter (wild-type (wt) and mutated) (Table S1) were dissolved into 10 mM potassium phosphate buffer (pH 7.0) or 1 × PBS (Phosphate buffered saline) (pH 7.4), in presence or absence of 100 mM potassium chloride (KCl). The sequences were annealed by heating at 95 °C for 5 min, followed by cooling slowly to room temperature to allow G4 formation. 1 × PBS was used to mimic physiological conditions for the in vitro studies.
Förster resonance energy transfer (FRET) studies
To understand the kinetics of G4 folding upon recombinant Nucleolin and Sp1 binding, we performed FRET studies, wherein the nonradiative energy is transferred from the excited donor fluorophore to the ground state acceptor fluorophore. To prepare the FRET probe, oligonucleotide sequences (GQ (wt) and C-wt (Table S1)) were procured from Eurofins India Pvt. Ltd, in which 6-FAM (Donor) was attached to the 5ꞌ-end of the G4 strand (GQ (wt)) and an internal Cy3 (Acceptor 1) tag was attached to 22T in the same strand. Cy5 (Acceptor 2) tag was attached to the complementary sequence of G4 strand (C-wt) at the 3′ -end. Two oligonucleotide sequences were allowed to form heteroduplex at an equimolar concentration by heating at 95 °C followed by slow cooling to room temperature. From steady-state emission intensity, the qualitative idea had been evaluated about the efficiency of energy transfer between the donor and acceptor fluorophores in presence of Nucleolin and Sp1. Oligonucleotide sequences were diluted to 1 µM into 1 × PBS (pH 7.4) while recombinant Nucleolin (0.25–3 µM) and Sp1 (0.25–1 µM) were titrated at increasing concentration gradient at 16 °C.
We acquired the fluorescence emission spectra from 560 to 690 nm wavelength by exciting the solution at 495 nm. Then, we determined the FRET efficiency and the distance between donor and acceptor fluorophores attached to the probe both in presence and absence of Nucleolin and Sp1 using the following equations:
Förster distance (R0) between 6-FAM and Cy3 pair is ~ 6.7 nm and 6-FAM and Cy5 is ~ 6.5 nm [41].
\({F}_{DA}\): Donor intensity in the presence of acceptor; \({F}_{D}\): Donor intensity in the absence of acceptor.
The distance (\({R}_{DA}\)) between 6-FAM—Cy3 and 6-FAM—Cy5 had been determined using the formula:
Dimethyl sulphate (DMS) protection assay
To analyze the composition of G-tetrad core and G4 folding topology, we performed DMS protection assay on 74-nucleotide long oligonucleotide sequence that encompasses two adjacent G4 sequences of MAPK12 promoter. The sequence was purchased from Xcelris Genomics Pvt. Ltd. and labelled with 6-FAM (6-Carboxyfluorescein) at 5ꞌ-end. We annealed the sequence by heating the samples at 90 °C for 5 min and then cooling slowly to room temperature in 20 μl of 10 mM potassium phosphate buffer (pH 7.0) with or without 100 mM KCl. We methylated the annealed samples by 0.1% (final concentration) DMS for 2 min at room temperature. The reaction was stopped by stop solution (0.3 M sodium acetate (pH 7.0) and 1 M β-mercaptoethanol), and subsequently ethanol-precipitated by adding 2.5X volume of chilled ethanol, 1.5X volume of 0.3 M sodium acetate (pH 5.2) and centrifugation at 12,000×g for 30 min at 4 °C. We washed the pellet with chilled 75% ethanol followed by centrifugation at 12,000×g for 30 min at 4 °C. We prepared the A + G ladder of the same 74-nucleotide long DNA fragment by adding 4% formic acid to 400 nM sequence following 30 min of incubation at 37 °C. We cleaved the methylated samples and the A + G ladder sample by 1 M piperidine for 20 min at 95 °C. The samples were further lyophilized to remove piperidine following three successive water washes. The cleaved products were dissolved in formamide dye (47.5% formamide, 0.01% SDS, 0.01% bromophenol blue, 0.005% xylene cyanol and 0.5 mM EDTA) and analyzed on a 15% sequencing (denaturing) urea-PAGE gel along with A + G sequencing ladder of the same 74-nucleotide long DNA fragment. Resolved DNA fragments were visualized using Typhoon Trio+ phosphorimager (GE Healthcare). Relative band intensity of each nucleotide fragment was calculated by dividing the intensity of target bands in the lane with the corresponding band in A + G ladder.
Biotin–streptavidin magnetic pull-down assay
To identify the proteins that bind to the G4-motifs at MAPK12 promoter, 5ꞌ-biotinylated wild-type G4 sequences were procured from Eurofins India Pvt. Ltd. and reconstituted into 1 × PBS (pH 7.4). We prepared the nuclear extract of MDAMB-231 cells from 70% confluent cells, in which cells were washed in 1 × PBS (pH 7.4), scraped, and pelleted down by centrifugation at 3500×g at room temperature for 5 min. The pellet was resuspended into nuclear extraction buffer (20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 0.4 mM NaCl, 0.2 mM EDTA, 25% glycerol, 0.5 mM PMSF, 0.5 mM DTT, and 0.1% Tween 20) and incubated for 1 h in ice. Cells were centrifuged at 12,000×g for 30 min at 4 °C to collect the supernatant with the nuclear extract. Dynabeads M-280 streptavidin were vigorously shaken in the preservative. 200 µl of beads per reaction were added to the microcentrifuge tube. The tube was secured on a Promega PolyATract 1000 magnet to pull down Dynabeads. The supernatant was carefully removed without disturbing the beads. Beads were washed by 500 µl of 2 × B/W buffer (10 mM Tris–Cl (pH 7.5), 1 mM EDTA, 2 M NaCl, 0.05% Tween 20) thrice. Following the final wash, beads were resuspended into 190 µl of 2 × B/W buffer without Tween 20, in which 200 µl of 250 ng/µl biotinylated G4 probe was added. The mixture was incubated for 20 min at room temperature under rocking condition. Then, beads were pulled down using the magnet and these steps were repeated thrice to saturate the beads with G4 probes. Probe-bead complex was washed in 400 µl of TE buffer three times. To make the probes suitable for protein–DNA interactions, the probe-beads were washed with 500 µl BS/THES buffer (22 mM Tris–Cl (pH 7.5), 4.4 mM EDTA, 8.9% sucrose (mass/vol), 62 mM NaCl, 0.3% protease inhibitory cocktail, 0.04% phosphatase inhibitory cocktail, 10 mM HEPES, 50 mM KCl, 5 mM CaCl2, 12% Glycerol), supplemented with 10 µg/ml Poly dI–dC to reduce nonspecific binding. To allow protein interaction to biotinylated G4, 200 µl BS/THES buffer was added to the probe–bead complex along with 750 µl of nuclear extract and incubated for 30 min at 4 °C under rocking condition. The bead–protein complex was pulled down using the magnet and the supernatant was discarded. The probe–bead–protein complex was further washed with 500 µl of BS/THES buffer with 10 µg/ml Poly dI–dC five times and in the final step, washing was done with 500 µl of BS/THES buffer without Poly dI–dC. The bound proteins were eluted in presence of 120 µl elution buffer (25 mM Tris–Cl (pH 7.0) and varying concentration of NaCl) at an increasing gradient of NaCl (50, 100, 250, and 350 mM). Elution fractions were run on 15% SDS-PAGE and proteins were detected by Western blot using anti-Nucleolin antibody (Abcam) and anti-Sp1 antibody (Sigma).
Cell culture and 3D spheroid assay
Human triple negative breast adenocarcinoma cell line, MDAMB-231 was procured from NCCS, Pune and was maintained in DMEM-complete media with 10% (v/v) FBS, 100 unit/ml penicillin, and 100 µg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2. For 3D culture (spheroid) of MDAMB-231 cells, cells were subcultured into DMEM media, supplemented with 20 ng/ml recombinant epidermal growth factor, B27 supplement, 5 μg/ml insulin, 0.4% BSA in a humidified atmosphere with 5% CO2 at 37 °C. Single-cell suspensions were prepared using mild enzymatic dissociation using 1 × Trypsin–EDTA solution at 37 °C for 2–3 min. To generate the spheroids of 300–400 µm diameter on the 4th day of inoculation, the dissociated cells were diluted from the stock cultures at a concentration of 7000 cells per ml in the standard medium. Then, 200 µl of cell suspension was transferred into each well of agarose-coated microtiter plate and incubated for 96 h in a humidified atmosphere with 5% CO2 at 37 °C. Spheroid formation was monitored by phase contrast microscope.
Dual-luciferase assay
We performed dual-luciferase assays to examine the intracellular selectivity of TMPyP4 and NDI compounds for MAPK12-G4. Dual-luciferase constructs of MAPK12 promoters with and without the G4 scaffolds were transfected into MDAMB-231 cells along with 50 ng of pGL3-control vectors (Promega) (used as internal control) using Lipofectamine® 2000 transfection reagent (Thermo-Fisher Scientific) following standard protocol recommended by the manufacturer. The role of compound-bound G4 complex in oncogene promoter activation was investigated in contrast to the G4-null promoter constructs (MAPK12 promoter cloned in luciferase construct without G4 motifs) into MDAMB-231 cells. Cells were sub-cultured into 24 well plates at a density of 2.5 × 104 cells per well and transfected with the reporter plasmids for 48 h. After 24 h of transfection, cells were treated with different concentrations of TMPyP4 and TGS24. 24 h post-treatment, cells were washed with 1 × PBS (pH 7.4) and scraped in ice using 1 × PLB (Passive lysis buffer). Luciferase activities were monitored by the dual-luciferase assay system (Promega) as per the manufacturer’s protocol. Luminescence of each sample is detected in GloMax® 20/20 Single-Tube Luminometer (Promega) in triplicates and averaged from three independent experiments.
Chromatin immunoprecipitation (ChIP)
ChIP assays were conducted to monitor the promoter occupancy of the transcription factors across MAPK12 and NANOG promoters in presence of the NDI compounds following standard protocols. ChIP-grade antibodies (Anti-Nucleolin antibody [4E2]-ChIP Grade (Abcam)), Anti Sp1 antibody (Sigma), Anti c-JUN Rabbit polyclonal antibody (Abcam), and anti-Hsp90β polyclonal antibody (Abcam)) were employed for the experiments. ChIP-isolated DNA was purified using MinElute Purification kit (Qiagen) and was amplified by PCR reactions using forward and reverse primers specific to the G4-enriched regions at MAPK12 or NANOG promoter (Table S2) with Phusion® High-Fidelity PCR Kit (NEB). Antirabbit IgG was employed for mock immunoprecipitation.
Co-immunoprecipitation
To examine physical interaction of HSP90, c-JUN, and MAPK12 with RAS, co-immunoprecipitation studies were carried out using Classic IP kit (Pierce). In brief, MDAMB-231 cells were grown at a density of 1 \(\times\) 105 cells per well and treated with 50 nM TGS24 for 24 h and were lysed in lysis buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, and the protease inhibitory cocktail. After centrifugation at 14,000 rpm for 30 min at 4℃, the supernatant was incubated with protein G or protein A/G-immobilized agarose beads (Santa Cruz), preloaded with anti-Ras antibody (Abcam). The immunocomplexes were extensively washed with the lysis buffer. Detection of HSP90, c-JUN, and MAPK12, immunoprecipitation with Rabbit monoclonal anti-Hsp90β antibody (Abcam), Anti c-JUN Rabbit polyclonal antibody (Abcam), Rabbit anti-p38γ-antibody (CST) in the IP buffer (20 mM Tris–HCl (pH 8.0), 0.2% Nonidet P-40, 1 mM dithiothreitol, and the protease inhibitory cocktail) followed by washing in the wash buffer that contains 1% Nonidet P-40. Resins were extensively washed, and the immune complexes were subsequently eluted and analysed by Western blotting with anti-Hsp90β or anti-c-JUN or anti-p38γ antibodies. The immunoprecipitants were transferred to the nitrocellulose membranes, which were immunoblotted with specific primary antibodies and then incubated with horseradish-peroxidase conjugated secondary antibody against mouse or rabbit immunoglobulin, followed by the detection with ECL (Electrochemiluminescence) immunoblotting detection method.
Next-generation sequencing
To analyze whole transcriptome expression profile upon 50 nM TGS24 treatment for 24 h, RNA-seq study was carried out in MDAMB-231 tumorspheres. Total RNA was extracted from the tumorspheres using TRIzol following standard protocol and RNA concentration and purity were estimated using spectrophotometer measurements (evaluating the ratio of absorbance at 260 nm and 280 nm as a measure of protein contamination, and the ratio of absorbance at 260 nm and 230 nm as a measure of carbohydrate contamination), and its integrity was examined using capillary electrophoresis (BioAnalyser 2100, Agilent Technologies, Santa Clara, CA) to ensure the achievement of RNA integrity number (RIN) (28S to 18S ribosomal RNA) > 9, required for the preparation of the libraries for next generation sequencing (NGS). Then, the total RNA was processed with RiboCop rRNA depletion kit (Lexogen) following manufacturer’s recommendation to remove ribosomal RNA from total RNA content.
NGS analyses of 50 nM TGS24-treated samples were performed at Medgenome India Pvt. Ltd. using Illumina® HiSeq 4000/x platform to obtain 150-bp paired-end reads. The sequencing depth for each sample was 100 million reads. The raw reads were filtered using Trimmomatic (v-0.36) for retaining high-quality sequence and adapter trimming. Other unwanted sequences, such as mitochondrial genome sequences, ribosomal RNAs, transfer RNAs, adapter sequences using Bowtie2 (2.2.4). Filtered reads were aligned to Human genome (hg19) using splice aware aligner like STAR to quantify reads mapped to each transcript. Total number of uniquely mapped read counts were estimated using FeatureCount (1.5.2). The raw read counts for control and treated samples were normalized using DESeq2. A distribution of these log2(fold change) values were found to be normally distributed, wherein the genes which were found to be 2 standard deviations away from the mean (mean + 2 × Standard deviation, mean- 2 × Standard deviation) were differential expressed. GO annotation and Reactome pathway information for differentially expressed genes were done using Panther database. RNA-seq data had been submitted to GEO database GSE168403 (Supporting information: Scheme 2 and Scheme 3). GSEA analysis have been performed to find out the top-ranking gene-sets and their differential expression upon TGS24 treatment.
Semi-quantitative and real-time PCR
To verify the results of RNA-seq analyses and to observe the effect of TGS24, semi-quantitative and real-time PCR experiments had been carried out in MDAMB-231 cells. RNA isolation, cDNA synthesis, and polymerase chain reactions were performed following the protocols. Housekeeping gene, GAPDH was used as an internal control. PCR primers were designed using Primer-BLAST, NCBI, and analyzed in OligoAnalyser 3.1-IDT (Table S3). The real-time PCR gene expression analysis was analyzed by ΔΔCt method and represented as fold change. The level of significance was calculated by paired t test. The experiments were performed with three technical replicates and two biological replicates.
Deletion of G4-forming region at MAPK12 promoter using CRISPR/Cas9 method
MAPK12 promoter-specific gRNA sequences were designed using a combination of prediction tools, such as gRNA scorer 1.0 and 2.0, CRISPRz, and GuideScan having zero predicted off-targets. The crRNA sequences were synthesized by Dharmacon. The bipartite synthetic gRNA had been heteroduplexed using equimolar concentrations of gene-specific crRNAs and a tracrRNA in 30 mM HEPES (pH 7.4), 100 mM potassium acetate at a final concentration of 1 µM. The heteroduplex was annealed by heating at 90 °C for 10 min followed by slowly cooling to reach the room temperature. Alt-R™ 3NLS Cas9 nuclease was diluted to 1 µM from a 10 mg/ml stock solution into Opti-MEM. To prepare a mixed solution of 60 nM ctRNP complex, 5.25 µl of 1 µM crRNA: tracrRNA complex was mixed with 1 µM diluted Cas9, on which 77 µl of Opti-MEM medium was added. The mixture was incubated at room temperature for 5 min. Then, 4.2 µl of Lipofectamine RNAiMAX (Thermo) was mixed with 83.3 µl of Opti-MEM, which was added to the ctRNP complex, diluting the ctRNP complex to 30 nM. Then, the ctRNP–lipid complexes were incubated at room temperature for 20 min and transfected into MDAMB-231 cells. After 48 h of transfection, genomic DNA was isolated, and the target region was PCR-amplified using specific primers. Deletion of the target region had been confirmed by Sanger sequencing PCR.
In vivo studies
4T1 (murine mammary carcinoma) tumor cells were acquired from the National Centre for Cell Science (NCCS) central cell repository, Pune and grown in RPMI 1640 media, containing 1 mM sodium pyruvate, nonessential amino acids, 2 mM L-glutamine, 100 units/l penicillin, 100 µg/ml streptomycin, 50 µg/ml gentamycin sulfate, 50 µg/ml plasmocure, and 10% FBS at 37 °C with 5% CO2. Female BalB/c mice of 6–8 weeks old were obtained from Centre for Translational Animal Research (CTAR), Bose Institute, Kolkata, India and maintained following the guiding principles of the animal ethical committee, as noted by CPCSEA guiding principles. Mice were acclimatized to the laboratory environment at 25 °C for 7 days under 12 h’ dark–light cycle, before the experiments. 4T1 cells (1.5 × 106 cells per animal) were injected in the mammary fat pad of the mice subcutaneously to grow palpable solid breast tumors. The animals were indiscriminately divided into one control and one treated group having 5 mice into each group. The control group 1 comprised of mice induced to grow breast tumor using 4T1 cells (1.5 × 106), injected intraperitoneally with 1 × PBS (pH 7.4). Once the tumors were palpable after 14th day of inoculation, the intra-peritoneal injection of compound TGS24 (800 pg per kg body weight) was initiated. The dose was given at an interval of 48 h for next 21 days. Animals were observed for clinical abnormalities twice each day to observe the clinical or toxicological symptoms. Mortality, if any, signs of toxicity, body weight, and food consumption were monitored for 21 days post-treatment. After 21 days, mice were sacrificed, and the tumors and major organs (liver and lungs) were collected and fixed in 10% formalin for histopathological analyses. Tumor volumes were calculated using the formula V = 1/2 (width2 × length) using caliper.
Nude mice were maintained at 22–25 ℃ temperatures in a highly sterile environment, and light–dark cycles of 12 h, while the mice are having access to pellet diet and drinking water. At the start of the experiment the female mice were 5 months old, with a body weight of around 20 g. MDAMB-231 cells were cultured using RPMI media, supplemented with 12% FBS. Around 3 lakh cells were diluted using 50 µl of 1X PBS and 50 µl of Matrigel (Corning) and instantly administered into each female nude mice with an intraperitoneal injection, in the breast region. Breast tumor growth was detected within 2 weeks and the treatment with TGS24 began with a dosage of 800 pg/kg body weight, injected intraperitoneally amid 48-h intervals, between each dose, with a total of four given dosages. The tumor tissues and major organs were isolated and tested for differential gene expression by qPCR from the RNA isolated from the tumor tissue, and histopathological analysis of the organs.
Results
Characterization of two putative G4 forming motifs identified at the promoter region of human MAPK12 gene
G-rich domains in the human genome endure topological constraints due to supercoiling stress, epigenetic modifications, oxidative damage, and transcription factors’ binding, which result in G4 formation in the double-helical platform [42]. In this study, we identified two G4 forming motifs separated by a 21 bp (base pair) spacer element, upstream of the transcription-start-site (TSS) at MAPK12 promoter (positions − 163 and − 204) (Fig. 1A). These motifs were found to be evolutionary conserved among the primates, as revealed from the sequence homology of G-rich domains in the phylogenetic groups (Figure S1). The two motifs were assigned the G-scores of 41 (GQ-1) and 42 (GQ-2) using QGRS (Quadruplex-forming G-Rich Sequences Mapper) mapper [43]. Since G4 formation destabilizes the proximal duplex regions [44], and the presence of two G4s in tandem position indicate complex gene regulatory activity on MAPK12 transcription [45, 46], the 74-nucleotide long wild-type sequence of MAPK12 promoter had been considered for the characterization of G4 structures in vitro. CD (Circular Dichroism) spectra of wild-type 74-nt sequence (GQ (wt)) revealed a positive maximum at 262 nm and a negative maximum at 242 nm, indicating the formation of parallel topology (Fig. 1B). CD thermal melting experiment showed that GQ (wt) folds into highly stable G4 structures in presence of KCl (melting temperature (Tm) = 79 ± 2.5 °C) (Fig. 1C), and its thermodynamic stability is further enhanced upon increasing KCl concentration (Fig. 1D). The molecularity of GQ (wt) was investigated by concentration dependent CD melting studies. The Tm values did not significantly change in the wild-type (GQ (wt)) and mutant (GQ(m1) and GQ(m2)) sequences upon increasing DNA concentration (Fig. 1E), suggesting that they are all folded in intramolecular conformations.
Identification of two putative G4-forming motifs at MAPK12 promoter. A Two adjacent G4-forming motifs, GQ-1 and GQ-2, mapped at −204 and −163 base pairs respectively upstream the transcription start site (TSS) of MAPK12 promoter, separated by 21 nt. B CD spectra of G4-forming motifs in presence of 100 mM KCl at pH 7.0 and 25 C temperature; GQ(wt), wild-type G4-forming motifs; GQ(m1), GQ-1 motif mutated; GQ(m2), GQ-2 motif mutated. C CD-melting curves of GQ(wt), GQ(m1), and GQ(m2) in presence of 100 mM KCl at pH 7.0 within a range of 10 C–95 °C temperature at 5 °C interval. Fraction of folded G4 calculated from the sigmoidal curves using the molar ellipticity values at 265 nm as a function of temperature. Error bars represent mean ± SE (N = 3). D KCl-dependency of G4-forming motifs; CD-Melting temperatures (Tm) of GQ(wt), GQ(m1), and GQ(m2) plotted against increasing concentrations of KCl at pH 7.0 and 25 °C. Error bars represent mean ± SE (N = 3). E Concentration-dependent G4-molecularity of GQ(wt), GQ(m1), and GQ(m2). CD-Melting temperatures (Tm) plotted against increasing concentrations of G4-DNA at pH 7.0 and 25 °C. Error bars represent mean ± SE (N = 3). F DMS protection assay using 74-nt long MAPK12 promoter sequence containing adjacently located GQ-1 and GQ-2 motifs in presence and absence of 100 mM KCl. Guanine residues in G-tetrads and loop elements are assigned based on the principle of DMS-mediated protection of N7 atoms in guanine bases required for G4-folding with respect to A + G ladder. Densitometric values defining the DMS-cleaved band intensities are provided in the bar plot. Error bars represent mean ± SE (N = 3). G2–G21 participate in GQ-1 formation; G44–G61 involved in GQ-2 folding. Red asterisks at G7 and G10 denote the propensity of guanines to occur in both tetrad core and loop elements. G Four predicted G4 topologies of GQ(wt) sequence at MAPK12 promoter determined by CD spectral profiles and DMS protection assays. Parallel G4 topologies of GQ-1 and GQ-2 with propeller loops (orange line) connected by a 21-nt spacer forming putative stem–loop structure
Specific guanine residues were systematically mutated in GQ-1 (GQ(m1)) or GQ-2 (GQ(m2)) motifs to disrupt G4 formation in either domain, for monitoring the effect of one G4 motif on the dynamics and stability of the other in tandem position. GQ(m1) showed no characteristic spectra of parallel G4 structure, compared to the wild-type sequence, and showed prominent decrease in thermal stability (Tm = 45 ± 3.2 °C) in the presence of KCl (Fig. 1B and C). However, mutations in GQ-2 (GQ(m2)) restored parallel G4 structure in GQ-1, as revealed from its CD spectra with higher thermal stability (Tm = 65 ± 2.2 °C), as compared to that of GQ(m1) (Fig. 1B and C). Although the thermodynamic stability of GQ(m2) was significantly decreased as compared to that of the wild-type sequence, its stability was enhanced upon increasing KCl concentration, suggesting higher propensity of GQ-1 to form G4 structures, compared to GQ-2 (Fig. 1D). This observation was further validated by the NMR (Nuclear Magnetic Resonance) studies showing broad envelope of the imino proton resonances between 10 to 12 ppm for GQ (wt) and GQ(m2) and disappearance of the same in GQ(m1) (Figure S2A). This finding is agreeable with the previous reports which claimed that adenines at the central loop regions reduce the stability and impart conformational dynamics to the G4 structures, because of stacking interactions with the guanines in the tetrads [47,48,49]. However, thymine residues in the loop regions are more favored to induce kinetic stability in the G4 structures, due to the differences in the solvation entropy and the geometric plasticity of thymine residues [50, 51]. The differences in the structural behavior of wild-type and mutant sequences in presence of potassium ions were precisely indicated by the numbers and linewidths of the imino signals in the 1H NMR spectra. The broad and poorly resolved peaks in GQ(wt) and GQ(m2) spectra, in the absence of KCl, envisaged heterogeneous mixtures of G4 structures, while in 100 mM KCl, the revelation of prominent imino signals of sharper linewidth in GQ (wt) and GQ(m2) indicated potassium-induced conformationally restricted structures (Figure S2B). Interestingly, we observed proton resonances at 13.2 ppm in both GQ (wt) and GQ(m2) in presence of KCl, having poor resolution and lower intensity, compared to that of the imino signals between 10 and 12 ppm, suggesting the formation of minor populations of Watson–Crick base pairs as well (Figure S2B). To further delineate the tetrad core structures and G4 folding topologies of GQ-1 and GQ-2, DMS protection assay was carried out for GQ (wt) sequence both in presence and absence of 100 mM KCl. In GQ-1 motif, DMS-cleaved products at G2, G3, G4, G8, G9, G13, G14, G15, and G19 positions which displayed lowest band intensities in presence of KCl, suggesting their participation in tetrad core. Interestingly, G7, G10, G18, and G21 showed DMS cleavage of higher intensities in presence of KCl, indicating their equal propensity to participate in the loop as well as in the tetrad core (Fig. 1F). Thus, the conformational dynamics and heterogeneous mixture of various G4 topologies within the wild-type sequence were quite evident. This observation was further confirmed by 1D 1H proton NMR spectra of 22-nt long GQ-1 sequence that harbored site-specific G → T mutations at G7, G10, G18, and G21 positions. Indeed, single G → T mutations at these positions restricted the conformational heterogeneity, as evident from the appearances of sharper imino signals. However, dual G → T mutations at G10 and G18 positions induced the most kinetically stable conformer with distinct and sharp twelve imino signals, indicating the conformationally stabilized predominant G4 arrangement in solution (Figure S2C). Similarly, the 1D 1H proton NMR spectra of 21-nt long GQ-2 sequence, having specific G → T mutations at positions G16 and G18, revealed the appearance of sharper imino signals due to further restriction of conformational dynamics and stabilization of a particular conformer (Figure S2D). These results suggested that both GQ-1 and GQ-2 can form G4 structures in solution, though GQ-1 forms a more thermodynamically favored and highly stabilized G4 structure as compared to GQ-2.
Combining the findings from CD spectra, NMR studies, and DMS protection assays, four plausible models of G4 structures of GQ (wt) have been proposed, as shown in Fig. 1G. In GQ-1, G2-G7/G8-G13-G18/G19 and G4-G9/G10-G15-G20/G21 formed the first and third quartets respectively, with respect to the 5ꞌ-end. The residues G7, G10, G18, and G21 retained the comparable possibilities to occur in the quartet plane as well as in the intervening propeller loops, concluded from the equivalent DMS footprinting band intensities of these guanines, in the absence and presence of potassium ions. Interestingly, the 21-nt long spacer element separating GQ-1 and GQ-2 motifs harbored several mirror-repeats, which may form Watson–Crick base pairing between G24:C34, C27:G31, and C28:G30 (also confirmed by the evolution of proton resonances at 13.2 ppm). In GQ-2, G59-G54-G49-G44, G60-G55-G50-G45, and G61-G56-G51-G46 constituted the first, second, and third quartets respectively, interconnected by propeller loops.
G4-targeting porphyrin and naphthalene diimide (NDI) derivative recognize and stabilize putative MAPK12-G4 motifs in vitro
To probe G4 formation within MAPK12 promoter, a porphyrin derivative, TMPyP4 (5,10,15,20-tetra(N-methyl-4-pyridyl) porphyrin) was primarily employed as a reference compound to assess its binding profiles with GQ (wt) and GQ-1 sequences. TMPyP4 has been studied extensively to bind to a variety of G4 structures containing diverse topological variations, and end-stacking interactions provide higher G4 to duplex selectivity in vitro [52, 53]. In this study, the fluorometric titration of GQ(wt) and GQ-1 sequences resulted in the significant increase in the emission intensity of TMPyP4 with a characteristic peak at 660 nm, a prominent depression at 690 nm, followed by a small hump at 720 nm (Figure S3A and S3B). A similar binding property had been previously observed upon G4 titrations into TMPyP4. π-stacking interaction between its porphyrin ring and the G-tetrad plane leads to the splitting of Q(0,0) and Q(0,1) bands and inhibits the free rotation of the N-methylpyridinium groups [54, 55]. Figure S3C depicted sigmoidal binding curves by plotting the relative increase in fluorescence intensities (λem = 665 nm) against G4 concentration suggesting high affinity binding between TMPyP4 and MAPK12-G4s having KD (Binding dissociation constant) values of 1.4 ± 0.05 µM and 2.3 ± 0.07 µM for GQ(wt) and GQ-1 motifs respectively. These observations were further supported by CD melting experiments, which showed that TMPyP4 introduced higher thermal stability in GQ(wt) and GQ-1 motifs in aqueous solution (ΔTm of GQ(wt) = 12 °C ± 3.1 and ΔTm of GQ-1 = 18 °C ± 0.76) (Figure S3D).
Due to TMPyP4-mediated promiscuous interactions to other biomolecular G4s and duplex [52, 53], and its interference with multiple signaling pathways [56,57,58], this compound had not been taken up for further studies. Previous investigation using naphthalene diimide (NDI) derivatives have been carried out by the Neidle group in details suggesting that they bind to parallel G4 structures with high affinity and exhibit anticancer properties, by selective targeting of G4 structures. Their effects are studied in the critical pathways related to pancreatic cancer. Some of the well-studied NDI derivatives include MM41 [59] and CM03 [60, 61]. These compounds are limited by their poor solvation and selectivity for G4 structures. Therefore, tetrasubstituted naphthalene diimide (NDI) derivative compounds, TGS24 (N,N'-Bis-(3-(1H-imidazol-1-yl)propyl)-2,6-di(3-(1H-imidazol-1-yl)propylamino)-1,4,5,8,-naphthalenetetracarboxylic diimide) and an isogenic compound TGS25 (N,N'-Bis-(3-(3-methyl-1H-imidazol-3-ium-1-yl)propyl)-2,6-di(3-(3-methyl-1H-imidazol-3-ium-1-yl)propyl)amino)-1,4,5,8-naphthalenetetracarboxylic diimide) was synthesized and used as the probes (Fig. 2A), to assess their binding selectivity for MAPK12-G4 motifs. Owing to planar electron-deficient aromatic core of NDI derivatives, they create quadrupole moment with partial negative charge, above and below the plane, and a partial positive charge around the periphery. Their chemical accessibility, the peculiar characteristics of their aromatic planar core, and the presence of protonable side arms had been exploited to provide π–π stacking interactions with the electron-rich G-tetrad showing higher duplex to G4 selectivity [62]. Furthermore, differential substitution of the side-arms in NDI compounds had been utilized to provide better selectivity for G4 scaffolds of divergent topologies [63]. The NDI core has been preserved and the side arms have been modified for TGS24, to aid in the interaction with the G4 loop regions. Compared to MM41, we can see that the methyl-groups have been replaced in the TGS24 molecule. The presence of tertiary amines aids in ready protonation, thus favoring the solvation state of the molecule (Fig. 2A and S4). In this study, the titrations of GQ(wt) sequence into TGS24 resulted in the sharp rise of absorbance at 640 and 660 nm respectively. MAPK12-G4 binding to TGS24 led to maximum hyperchromic shift in TGS24 UV absorbance spectrum compared to other oncogenic G4 structures (e.g., G4 structures at BCL-2, VEGF, KRAS, and telomere (Tel22)) (Figure S5A), suggesting that TGS24 interacts with many G4 structures with different affinities. Since the UV absorbance spectrum for c-MYC G4 almost completely overlapped with the MAPK12 G4 spectra in presence of TGS24, we sought to calculate the apparent Kd (Kd, app) values (Figure S5B and S5C). Although the Kd value for MAPK12 was not significantly higher than that of c-MYC G4, which was also expected given the similarity in their G4 sequences, and the similar changes in their absorbance spectrum upon TGS24 binding, TGS24 showed slightly higher affinity for MAPK12-G4 (Figure S5D). Also, in fluorometric titration, we observed rise in the emission intensity and the Job’s plot suggested a 1:1 binding stoichiometry (Figure S6A and S6B). In aqueous solution, highly polar water molecules facilitate intramolecular charge transfer in the chromophore moieties of NDI compound, thus lowering the energy of the first singlet excited state and alleviates the triplet state transition which causes fluorescence quenching [64]. Unlike TMPyP4, TGS24 showed higher affinity for parallel G4s and showed better duplex-G4 selectivity, which was also confirmed by the Stephen Neidle’s group [65]. Since the fluorescent property of NDI compound was highly reduced in aqueous solution, we also performed ITC (Isothermal Titration Calorimetry) experiments to estimate the thermodynamic parameters of G4-binding reactions. TGS24 also showed higher affinity interaction with GQ(wt) as compared to the isogenic compound, TGS25 (Fig. 2B). Furthermore, TGS24 was found to highly stabilize the MAPK12-G4 structures compared to the duplex. TGS24 provided greater thermal stability to GQ(wt) (ΔTm = 18 °C ± 0.81) and GQ-1 (ΔTm = 20 °C ± 0.81) in aqueous solution (Figure S3E). These findings collectively suggested that TGS24 provided higher selectivity to the G4 motifs at MAPK12 promoter, that strongly escalated the thermodynamic stability of MAPK12-G4 structures. While TGS24 did not exhibit remarkable selectivity for MAPK12-G4, it displayed a higher level of specificity as compared to both TMPyP4 and the isogenic compound TGS25. As a result, we decided to conduct preliminary cellular studies by employing both TGS25 and TGS24 molecules. TGS25, being relatively nonspecific, was included in our investigations to provide a basis of comparison with TGS24, which showed greater specificity towards MAPK12-G4. This approach enabled us to assess and compare the downstream effects of targeting MAPK12-G4 with TGS25 as a reference. However, it is crucial to acknowledge that, at this stage, we cannot definitively ascertain whether TGS24 exclusively interacts with other promoter G4s. While our research predominantly provides valuable insights into the correlation between TGS24, MAPK12 expression, and the identified promoter G4s, it remains an open question whether the molecule engages with other G4 structures in the genome. This limitation underscores the need for further investigations to comprehensively understand the specificity and interactions of TGS24 with G-quadruplex structures.
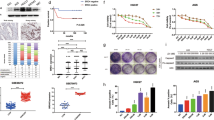
MAPK12-G4 binding selectivity and anticancer properties of naphthalene diimide compound. A Chemical formula of synthetic naphthalene diimide derivatives, TGS24 and another isogenic compound TGS25. B ITC profiles showing intermolecular interactions between wild-type MAPK12-GQ and TGS24, TGS25 in potassium phosphate buffer (pH 7.0) and 100 mM KCl at 25 °C temperature. Top panels: enthalpic heat released versus time at 25 °C during titrations. Bottom panels: thermogram of the integrated peak intensities plotted against the molar ratio of the complex. Thermodynamic parameters of intermolecular interactions (change in binding enthalpy (ΔH), entropy (ΔS), binding stoichiometry (n) and binding free energy (ΔG) are estimated from best-fit curves. C Flow cytometric analysis of apoptosis in MDAMB-231 cells by Annexin V-FITC-PI dual staining assay. 2 × 105 cells treated with TGS24 (50 nM and 100 nM) for 24 h. FITC: Fluorescein isothiocyanate; PI: Propidium iodide. In the typical flow cytometric quadrants, lower left quadrant (Annexin V-FITC negative/PI negative) denotes live cells, lower right quadrant (Annexin V-FITC positive/PI negative) denotes early apoptotic cells, upper quadrants (Annexin V-FITC positive/PI positive and Annexin V-FITC negative/PI positive) define late apoptotic and dead cells. D Percentage of live (Annexin V-FITC negative/PI negative), early apoptotic (Annexin V-FITC positive/PI negative), and late apoptotic/dead (Annexin V-FITC positive/PI positive and Annexin V-FITC negative/PI positive) cells calculated from flow cytometric analyses in MDAMB-231 cells treated with TGS24 (50 nM and 100 nM) for 24 h. Error bars represent mean ± SE (N = 3). E pGL4.72[hRlucCP] luciferase constructs having the inserts containing MAPK12 promoter sequences having both of wild-type G4-elements (wt), single-deletion of GQ-1 motif (GQ(d1)) or GQ-2 motif (GQ(d2)), and double-deletions of GQ-1 and GQ-2 (GQ-null) ahead of the hRluc coding region. The promoter sequences are cloned into KpnI and HindIII restriction sites. hRluc, Renilla luciferase gene; P1, promoter sequences. F Dual-luciferase assays. Evaluation of MAPK12 promoter activities using the luciferase constructs with or without the wild-type G4-forming sequences in MDAMB-231 cells. Promoter activities are determined by Renilla/Firefly luminescence values in control cells and cells treated with TGS24 (50 nM and 100 nM) and TGS25 (1 and 2 μM) for 24 h. Error bars represent mean ± SE (N = 3). Statistical differences in the luciferase activities compared to that of the control cells use two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). G Tumor regression by TGS24 (800 pg per kg body weight) treatment and without treatment (H) in BALB/c mice (average body weight 25 g) bearing palpable 4T1-breast tumors. Tumor volume measured up to 21 days at a 5 days interval after tumor implantation. Statistical analyses of tumor regression compared to that of the 0th day post-TGS24 treatment use two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001)
TGS24 exerts anticancer properties by downregulating G4-driven MAPK12 promoter activity and sensitizes MDAMB-231 cells to apoptotic death
Previously, NDI scaffold has been widely used to develop G4-binding molecules possessing in vivo anticancer activities, of which a few had reached the clinical trials phase [59, 66,67,68,69]. In this study, TGS24 had been taken up for the evaluation of anticancer activities and target selectivity for MAPK12-G4 motifs in MDAMB-231 cells. MTT assay showed that TGS24 exerted antineoplastic properties having IC50 values within a range of 34–64 nM in various cancer cell lines. (Table S4). Previous reports showed that MAPK12 overexpression promotes tumorigenesis and is one of the salient properties of triple-negative breast cancer (TNBC) cells [6, 70, 71]. Therefore, MDAMB-231 cell line had been chosen as a model for TNBC cells. Flow cytometric analysis using Annexin-V-FITC/PI staining suggested that TGS24 sensitized MDAMB-231 breast cancer cells to apoptotic death (Fig. 2C and D) at 50 nM concentration, as evident by the quantitative estimation of live, early apoptotic, and necrotic cell population (Fig. 2D). TGS24-driven increased expression of CASPASE 8, APAF-1, and cleaved PARP-1 was observed in MDAMB-231 cells (Figure S7A). In addition, accumulated release of CASPASE 3 was observed upon TGS24 treatment, which confirmed its potential to induce apoptotic signaling at a significantly lower concentration (Figure S7B and S7C). The binding specificity and promoter activity of MAPK12-G4 structures in the presence of TGS24 (and TGS25), in cellular microenvironment, was assessed by dual-luciferase assays. MDAMB-231 cells were transfected with the respective luciferase constructs (cloned in pGL4.72[hRlucCP]) containing the MAPK12-G4 motifs upstream of the Renilla luciferase coding sequences. The quantification of MAPK12 promoter activity with increasing concentrations of TGS24 was carried out, considering that the stabilization of G4 structure by the molecule would downregulate the expression of Renilla luciferase gene in contrast to the GQ-null plasmids and the plasmids carrying deletion of GQ-1 (GQ(d1)) or GQ-2 (GQ(d2)) motifs (Fig. 2E and F). The luciferase deletion constructs GQ-1 and GQ-2 motifs (GQ-null) showed constitutive activation of promoter even after TGS24 and TGS25 treatment, while the constructs with wild-type MAPK12-G4 motifs manifested ~ 70% downregulation of promoter activity, compared to that of GQ-null. This inhibition was more pronounced upon dose dependent TGS24 and TGS25 treatment. (Fig. 2E and F). This demonstrated the stabilization of G4 motifs by TGS24 and TGS25 have strong inhibitory effect on MAPK12 promoter activation and TGS24 and TGS25 could target and stabilize these motifs. Deletion of GQ-2 or GQ(d2) constructs showed inhibitory effect in promoter activation upon TGS24 and TGS25 treatment, comparable to wild-type (wt) construct. However, GQ-1 deletion or GQ(d1) upregulated the promoter activity compared to the wild type, which further supports the higher propensity of GQ-1 to form G4 structures, compared to GQ-2; TGS24 failed to reduce the promoter activation in GQ(d1), while TGS25 exhibited inhibition of promoter activity in GQ(d1), suggesting its lower selectivity for MAPK12-G4 motifs (Fig. 2E and F). Therefore, considering the binding selectivity and anticancer properties, TGS24 seemed to be the better candidate, compared to TGS25.
Anticancer activity of TGS24 was further evidenced in the in vivo studies in Balb/c female mice. Subcutaneous injection of 4T1 breast cancer cells (TNBC cells) in the left mammary pad of Balb/c mice was successfully used to develop palpable breast tumors, noticeable from the 14th day. Intraperitoneal dose of 800 pg/kg body weight of TGS24 resulted in ~ 80% regression of tumor size in 21 days, compared to the placebo, without significant changes in body weight (Figure S8A,S8B, Fig. 2G and H). Histopathological studies of the tumor and major organs, such as livers and lungs showed widespread metastases in the placebo group of mice, characterized by the enrichment and invasion of irregular large cell nuclei with multiple nucleoli. Treatment of TGS24 (800 pg/kg body weight) for 21 days resulted in the clearance of the tumor cells, as revealed from the loosely arranged cells in the microscopic images (Figure S8C). Here we adjusted the in vivo dosage based on the IC50 values obtained from our cell line studies. We focused on optimizing the therapeutic outcome while minimizing potential injury to the animals. The chosen dose showcased promising results, and our rationale was guided by a conscientious approach that prioritized the animals' well-being while achieving meaningful tumor reduction.
TGS24 exerts inhibition in cell migration and cancer stem cell expansion in MDAMB-231 (TNBC) cells and xenografted nude mice model
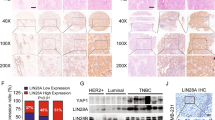
Nuclear localization of TGS24 in MDAMB-231 cells was clearly visible from the confocal microscopic images after 12 and 24 h of treatment (Figure S9A). Based on the mean fluorescence intensity, nuclear uptake efficiency of TGS24 was significant (Figure S9B), supporting the idea that TGS24-induced stronger cellular responses in apoptosis and MAPK12 promoter inhibition. Generation of MAPK12-knockout mice has recently provided ground-breaking information regarding the innumerable functions of MAPK12, which includes its role in stemness and metastasis regulation [4, 8, 18, 21, 23, 72, 73]. Therefore, the sphere forming efficiency (SFE) had been assessed in MDAMB-231 cells upon TGS24 treatment to observe its effects in stemness regulation. The microscopic images showed decreased numbers and size of tumorspheres in MDAMB-231 cells (Figure S9C). The SFE of MDAMB-231 cells in sphere culture was suppressed by ~ 60% with TGS24 treatment at a significantly lower concentration (Figure S9D). In addition, TGS24 weakened the migratory potential of MDAMB-231 cells by ~ 40% at 24 h (compared to the control), as revealed from the wound-healing assay (Figure S9E and S9F). Consistent with the previous results of apoptosis induction and MAPK12 selectivity, coupled with the wound-healing assay and tumorsphere forming studies, TGS24 was found to be a promising therapeutic candidate. Next, TNBC cells were xenografted in nude mice to develop tumor in the mammary region. The tumors were developed within 2 weeks of xenografting, and the nude mice were treated with TGS24 at a dosage of 800 ng/kg body weight. The tumors and the vital organs (like liver, kidney, lungs) were isolated and used for histopathology (Fig. 3A), which showed that the metastatic malignant cells are reduced in TGS24 treated condition (Fig. 3B). The tumor tissue was used for RNA isolation and subsequent analysis of gene expression against GAPDH control. The stemness related genes (OCT4, SOX2, NANOG) all showcased decrease in expression by the comparative ΔΔCt method. The expression of MAPK12, RAS, CD44, cMYC, HSP90, cJUN, VEGF and ZEB1 was also decreased significantly (Fig. 3C). Indeed, the stemness properties of the cells are evidently lost after TGS24 treatment in nude mice, along with downregulation of genes responsible for cancer cell invasiveness and metastasis.
Xenograft model of TNBC cell line MDAMB-231 in NUDE mice. A Schematics of xenografting procedure to establish tumor in NUDE mice and subsequent dissection procedures for histopathological and gene expression analysis. B Histopathological analysis of the prepared tissue sections of the major organs like kidney, liver and lungs by haematoxylin eosin staining and fixed slides imaged using inverted phase-contrast microscope. The major organs are isolated from TGS24 treated mouse and compared with the no treatment control group. C Gene expression analysis by real-time PCR for the various genes promoting stemness, metastasis and key drivers of oncogenesis. RNA isolated from the tumor tissue of xenografted mice model is used for the experiment. Relative gene expression with respect to GAPDH housekeeping gene. (*P < 0.05, **P < 0.01)
Binding of Nucleolin and Sp1 at G4 motifs within the MAPK12 promoter
Two adjacently located G4 motifs have been identified to inhibit MAPK12 transcription, while mutations in G4 scaffold enhanced MAPK12 promoter activation. Hence, it is speculated that specific transcription factors regulate the kinetics of G4 folding–unfolding process to modulate MAPK12 transcription. Previous reports have shown that Sp1 increases MAPK12 transcription through binding to the G-box elements present in its promoter region [39]. A recent study by Sutherland et al. depicted that Sp1 binding to the G-rich domains increases negative super helical stress, that inhibits Nucleolin occupancy in c-MYC-G4 element [74]. Since Sp1 and Nucleolin bind to various G4 structures across the genome and modulate G4-mediated transcription of many oncogenes [75,76,77], we performed western blots using Sp1- and Nucleolin-specific antibodies following biotin-streptavidin magnetic pull-down of MAPK12-G4 elements. In this study, biotin labelled G4 motifs in MAPK12 promoter have been probed to identify if Nucleolin and Sp1 interact with the G4 structures. Biotin-conjugated G4 motifs were attached to the streptavidin-coated magnetic beads and incubated in the nuclear extract of MDAMB-231, to allow the binding of specific transcription factors. The bound transcription factors were eluted with an increasing gradient of NaCl followed by subsequent detection with Nucleolin- or Sp1-specific antibodies in western blot experiments (Figure S10A). This suggested the binding of Nucleolin and Sp1 proteins to the G4-binding domains (Figure S10B). To verify this observation, we performed ChIP experiments on the region encompassing adjacent G4 elements within the MAPK12 promoter and estimated the efficiency of Nucleolin and Sp1 binding in MDAMB-231 cells. We also performed the ChIP experiment under siRNA-knockdown of Nucleolin to investigate if Nucleolin knockdown affects Sp1 binding. However, their relationship does not seem to be reciprocally related, nor do they compete for binding because the increase in Sp1 occupancy is not statistically significant compared to the control, and the error bars overlap. This observation aligned with the structural complexity of the MAPK12-G4, which spans a considerable length and may not possess overlapping binding sites for Sp1 and Nucleolin. Furthermore, our findings suggest that there may be other transcription factors at play in the intricate regulation of MAPK12 expression. The binding of Nucleolin, which stabilizes the G4, and Sp1, which appears to have a destabilizing effect implies the involvement of a more complex regulatory network (Figure S10C and S10D). To confirm if the neighboring G4 elements are target binding sites of Nucleolin and Sp1 at MAPK12 promoter, we employed CRISPR/Cas9-mediated deletion of the G4-motifs at MAPK12 promoter, which revealed strong reduction of both Nucleolin and Sp1 recruitment, suggesting their specific interactions with the G4 elements in the promoter region. We first attempted to introduce point mutations using CRISPR/Cas9; however, due to high G-richness in the MAPK12-G4 domains, it resulted in numerous -specific effects. Therefore, we introduced deletion to successfully remove the two G4 forming motifs from the promoter. Although removing a long stretch of sequence from the promoter region might inadvertently remove some important domains, it also encompassed the G4 motifs, which have been found to be indispensable to regulate its transcription. However, due to the potential nonspecific biological effects of the deletion of two G4-motifs and the 21 bp intermediate region, we chose to study the effect of MAPK12-G4s upon TGS24 treatment, as it presented a better candidate to target MAPK12-G4s, as shown by both in vitro and luciferase studies.
Nucleolin and Sp1 regulate duplex-to-G4 kinetics to poise MAPK12 transcription homeostasis
Studies have, so far, suggested that GQ-1 shares the common binding site for both Nucleolin and Sp1 and exhibits greater propensity to fold into G4 structure, as compared to GQ-2. Therefore, to figure out Nucleolin and Sp1-induced G4 plasticity at GQ-1 and their role in regulating the kinetics of its folding-unfolding process, we carried out FRET studies using a duplex nucleotide probe that hosts the GC-rich sequences of MAPK12 promoter, having 6-FAM at the 5ꞌ-end of GQ-1 and an internal fluorophore, Cy3 at T22 within the same strand. G4-hosting strand was hybridized with its complementary strand that contained Cy5 tag at the 3ꞌ-end (Fig. 4A). Since the complementary strand of G4-hosting sequence had a high proportion of cytosine residues, which might give rise to i-motif formation, CD spectroscopy was performed to determine the secondary structure of the C-rich strand alone and in hybridized form with the G-rich strand. C-rich strand upon hybridization with the G4-hosting sequence resulted in duplex formation under pH 7.0 and pH 3.0, as revealed by the positive maxima at 275 nm. In absence of the G-rich strand, under acidic conditions (pH 3.0), positive maxima at 285 nm and negative maxima at 258 nm clearly demonstrated the formation of i-motif structure in the C-rich strand. However, in the presence of the G-rich strand, positive maxima at 275 nm suggested the predominant formation of duplex under pH 7.0 and pH 3.0 at 25 °C. In the presence of 100 mM KCl, positive maxima at 265 nm and a shoulder around 275 nm, suggested the co-existence of both duplex and G4 structures (Figure S11). These CD spectra collectively indicated that the GC-rich duplex at MAPK12 promoter involves an equilibrium between G4 and duplex under physiological pH. Fluorometric titration of Nucleolin resulted in high FRET signal between 6-FAM and Cy3 suggesting Nucleolin-induced G4 formation at GQ-1. The gradual decrease in the FRET signal between 6-FAM and Cy5, indicated the disruption of duplex formation (Fig. 4A and B). In contrast, fluorometric titration of Sp1 led to strong FRET signal between 6-FAM and Cy5, indicating Sp1-induced duplex formation. Concomitantly, Sp1 titration gradually reduced the intensity of FRET signal between 6-FAM and Cy3, indicating Sp1-induced G4 resolution (Fig. 4A and C). With increasing Sp1 concentration, FRET efficiency between 6-FAM and Cy5 was increased by ~ 20%, while an increase in Nucleolin decreased the FRET efficiency by ~ 30% (Fig. 4D). Since donor fluorescence quantum yield has a direct influence on the Forster distance (R0) and FRET efficiencies, the quenching effect was taken into consideration to estimate the change of dye-to-dye distance upon protein titrations using the following equation: E = R06/(R06 + r6). The distance between 6-FAM and Cy3 decreased from 6.5 ± 0.1 nm (R0 = 6.5 nm) in the free DNA to 5.2 ± 0.1 nm in the complex with Nucleolin, indicating G4 formation. The distance between 6-FAM and Cy5 decreased from 6.8 ± 0.2 in free DNA to 5.3 ± 0.3 under Sp1-bound conditions, indicating duplex formation. Since Nucleolin and Sp1 regulate the duplex-to-G4 kinetics at the MAPK12 promoter, we performed dual-luciferase assays to investigate the effect of these proteins on G4-driven MAPK12 promoter activation. siRNA-knockdown of Nucleolin strongly activated MAPK12 promoter activity, while TGS24 treatment resulted in decreased promoter activity and transcription of the MAPK12 gene (Fig. 4E and F). To examine the gene-regulatory functions of dual G4 elements, we employed CRISPR/Cas9-mediated disruption of G4 motifs resulting in undetectable transcripts’ level of MAPK12, which led to significant growth arrest and cellular death. CRISPR/Cas9-mediated disruption of G4 elements underscored the role of G4 elements to poise homeostatic regulation of MAPK12 transcription. The duplex-to-G4 equilibria at neighboring G4 motifs and binding of Nucleolin or Sp1 are pivotal in maintaining basal level of MAPK12 expression within the cells. Furthermore, the observation that TGS24 (50 nM) treatment decreased the promoter activation, confirmed the biological significance of G4 motifs. Displacement of Nucleolin and Sp1 by TGS24 from G4 structures appended bona fide stability to the G4 motifs, resulting in the inhibition of MAPK12 promoter activation (Fig. 4E). This was in accord with MAPK12 transcription reduction by TGS24 (50 nM) treatment (Fig. 4F).
Nucleolin and Sp1 regulate G4 → duplex transition at MAPK12 promoter. A FRET probe. MAPK12-G4 motifs conjugated with 6-FAM at 5′-end of GQ-1 and internal Cy3 at T22. The complementary strand of G4-forming sequence contains Cy5 at 3′-end. G4-formation gives high FRET signal between 6-FAM and Cy3. Duplex formation gives high FRET signal between 6-FAM and Cy5. B Fluorometric titration of 90 nM FRET probe with increasing gradient of recombinant Nucleolin (0–3 µM) in 50 mM Tris–Cl, pH 7.0, 100 mM KCl at 15 °C at excitation wavelength (λex) of 497 nm. Decline of FRET signal at Cy5 emission ((λem) 650 nm) upon Nucleolin titration suggests disruption of duplex with concomitant increase of Cy3 emission ((λem) 595 nm) indicating G4 evolution. C Fluorometric titration of 90 nM FRET probe with increasing gradient of recombinant Sp1 (0–1 µM) in 50 mM Tris–Cl, pH 7.0, 100 mM KCl at 15 °C at excitation wavelength (λex) of 497 nm. Decline of FRET signal at Cy3 emission ((λem) 595 nm) upon Sp1 titration suggests G4 resolution with concomitant increase of Cy5 emission ((λem) 650 nm) indicating duplex transition. D FRET efficiency calculated following the equation: E = 1 – (IDA/ID); IDA and ID are the emission intensity of donor fluorophore in presence and absence of acceptor fluorophore respectively. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). With increasing Sp1, distance between 6-FAM and Cy5 decreases. Nucleolin titration decreases the distance between 6-FAM and Cy3. The change in distances (r) between donor and acceptor fluorophore calculated by E = R06/(R06 + r6). R0 denotes Forster distance between donor and acceptor. The distance between 6-FAM and Cy3 decreased from 6.5 ± 0.1 nm (R0 = 6.5 nm) in the free DNA to 5.2 ± 0.1 nm in the complex with Nucleolin indicating G4 formation. The distance between 6-FAM and Cy5 decreased from 6.8 ± 0.2 in free DNA to 5.3 ± 0.3 under Sp1-bound conditions indicating duplex formation (E) pGL4.72[hRlucCP] luciferase constructs having the inserts containing MAPK12 promoter sequences having adjacent G4-elements (wild-type). Dual-luciferase assays. Evaluation of MAPK12 promoter activities in MDAMB-231 cells under the following conditions: siRNA knockdown of Nucleolin at 48 h and TGS24 (50 nM) treatment for 24 h, nucleolin-knockdown + TGS24 treatment, nucleolin knockdown + TGS25 treatment, and TGS24 and TGS25 treated alone separately Error bars represent mean ± SE (N = 3). Statistical differences in the luciferase activities compared to that of the control cells use two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). Statistical differences in the luciferase activities in presence of TGS24-treated samples compared to that of cells, treated with recombinant Sp1 and Nucleolin, siRNA-knockdown of Nucleolin use two-tailed Student's t test (###P < 0.001). F RT-PCR analysis. Expression profile of MAPK12 transcripts from P1 promoter in MDAMB-231 cells under following conditions: TGS24 (50 nM) treatment for 24 h and siRNA knockdown of Nucleolin for 48 h, nucleolin-knockdown + TGS24 treatment, nucleolin knockdown + TGS25 treatment, and TGS25 treated alone separately. Quantification of the transcripts’ level relative to the control by qPCR analyses. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). Statistical differences between recombinant Nucleolin-treated and Nucleolin-knockdown cells are determined by two-tailed Student's t test (###P < 0.001). GAPDH considered as housekeeping gene
Downregulation of G4-driven MAPK12 expression by TGS24 abrogates stemness expansion, and metastatic potential of cancer cells and triggers apoptotic signaling
To perform comprehensive transcriptomic analysis of MDAMB-231 cells upon TGS24 (50 nM) treatment for 24 h, differential mRNA expression of the oncogenes have been estimated using next-generation sequencing (NGS) approach. We chose the time-point based on our previous cellular experiments that allowed us to observe the effects of the compound on MAPK12 expression, without inducing complete cell death. It presented us an ideal window for assessing the impact of the compound. The study optimized an RNA-seq pipeline to map the read counts to human transcriptome and compare the gene expressions in TGS24-treated cells with respect to the untreated ones. NGS data revealed that among 1321 total genes detected 269 were differentially upregulated, while 346 were differentially downregulated with statistical significance (FDR > 0.05, and fold change > 2), in the TGS24-treated cells as compared to the untreated ones. Of them, 44 top-scoring genes are associated with metastasis, stemness, proliferation, and apoptosis-related pathways, revealed distinct on/off switch. This suggested a pattern of upregulated or downregulated genes with respect to the differential expression of MAPK12. Of particular significance within this set of genes are PARP1, NANOG, and hnRNP K. These genes are reported to harbor distinct G4 structures located within their promoter regions. Notably, the nature of these G4 formations is not uniform. In the case of PARP1, the G4 structure adopts a hybrid configuration [78], while the G4 region within the hnRNP K promoter displays a mixed arrangement, encompassing both G4 and i-motif structures [79]. While a promoter G4 structure had been observed in NANOG in the sequencing study by Lyu et al. in a recent article [80], G4 formation or its potential functions are not extensively investigated. It is important to acknowledge that among the genes showing differential expression in the RNA-seq study, there is a possibility that some may host G4-forming domains. These G4 structures, if present, might not have been extensively studied or characterized to date. Intriguingly, our findings align harmoniously with prior research conducted by the Stephen Neidle's group [65], corroborating the heightened binding preference of TGS24 for parallel G4 structures. Therefore, the differential expression of the oncogenes, reported in this study may be a combined effect of MAPK12-G4 targeting and nonspecific binding of TGS24 to other putative G4s in the genome [80, 81]. However, as a pivotal player operating at the crossroads of stemness, apoptosis, and metastasis pathways, MAPK12 exerts regulatory control over numerous downstream genes. Consequently, the observed alterations in the expression of these genes might also stem from the intricate network of interactions orchestrated by MAPK12. Vimentin, MMP12, CD44, and SOX2, associated with metastasis and stemness pathways were differentially downregulated upon TGS24 treatment, suggesting that reduced expression of these oncogenes might lead to diminution of migratory potential and sphere forming rates in MDAMB-231 cells. However, Wnt, β-Catenin, and ALDHβ1 expression remained unaltered upon TGS24 treatment (Fig. 5A and C). Furthermore, downregulation of RAS, c-FOS, c-JUN, and MAPK12 upon TGS24 treatment indicated that silencing of MAPK12 expression by TGS24 reversed the expression of these proliferation markers of RAS pathway, leading to the reduced oncogenic transformation of RAS (Fig. 5A and B). Furthermore, enhanced expression of GATA3, CASPASE 9, and PARP-1 and reduced expression of NF-κB suggested that TGS24 treatment led to the increase of pro-apoptotic markers, sensitizing the cells to apoptotic death (Fig. 5A and D). To validate NGS analyses, both semi-quantitative and real-time PCR experiment have been performed on the selected genes, such as RAS, MAPK12, c-FOS, Vimentin, MMP12, NF-κβ, c-JUN, SOX2, and CD44 which showed robust downregulation in TGS24-treated cells, as compared to the untreated ones (Fig. 5B–D). We sought to enhance the robustness of our findings by comparing our RNA-seq data with a dataset previously generated by Neidle's group [60, 82]. Both studies employed similar NDI compounds to explore their potential effects on cancer cell lines. This comparative analysis provided valuable insights into the consistency and reproducibility of the observed transcriptional alterations. Strikingly, the RNA-seq data from both studies independently underscored the perturbation of crucial signaling pathways, including RAS, MAPK, Wnt, and Notch, upon treatment with NDI compounds. This alignment reinforces the significance of these pathways as key players in the cellular responses to NDI treatment. Importantly, our dataset further accentuates the significance of MAPK12 within this context. Building upon Neidle's group's observation, we discovered MAPK12 to be one of the major targets influenced by NDI treatment. The convergence of our findings on RAS and stem cell-related pathways from both datasets provides compelling evidence, reinforcing the understanding of the compound's impact on cancer cells. The results of linear regression analysis further indicated a high correlation (R2 = 0.854) between the data of RT-qPCR and NGS analyses (Fig. 5E). This observation further suggested that MAPK12-downregulation led to the reduction of stemness and metastatic markers in cancer cells, which was reflected in reduced sphere formation and the inhibition of migratory properties of MDAMB-231 cells (Figure S12). TGS24 also reduced the expression of oncogenes, related to RAS pathway leading to the antiproliferation of cancer cells and subsequent sensitization of cancer cells to apoptosis. This finding was consistent with the results of flow cytometry. Therefore, MAPK12-induced stemness and metastatic pathways might be at the crossroad of RAS pathway, wherein MAPK12 might act as the essential effector protein connecting the crosstalk between RAS-mediated proliferation, stemness and metastasis-related cascades (Fig. 5F, S12).
Downstream effects of TGS24-treatment and its selective interaction with MAPK12-G4 motifs. A RNAseq analyses of TGS24 treatment in MDAMB-231 spheres expressing high MAPK12. Heatmap of biological replicates of differentially expressed genes upon 24 h treatment of 50 nM TGS24 treatment in MDAMB-231 spheres using the log10(count) values for each replicate. The screening threshold for the differentially expressed genes is set to: |log2(Fold Change)|> 1 and P value < 0.05. Differential expression of the transcripts clustered by Euclidean correlation, with distinct upregulation, unaltered, and downregulation patterns in expression for TGS24 treatment, compared to control condition. The count values are colour coded blue to yellow to red in increasing order. Differentially expressed genes are clustered based on their involvement with RAS, apoptosis, and stemness- and metastasis-related pathways. B RT-qPCR-verification of RNAseq data for differentially expressed genes in RAS pathway upon 50 nM TGS24 treatment in MDAMB-231 spheres for 24 h: MAP4K3, RAS, MAPK12, c-FOS, c-JUN, HSP90. Quantification of the transcripts’ level relative to the control by qPCR analyses. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). GAPDH considered as housekeeping gene. C RT-qPCR-verification of RNAseq data for differentially expressed genes in metastasis and stemness-related pathway upon 50 nM TGS24 treatment in MDAMB-231 spheres for 24 h: ALDHβ1, WNT-1, β-Catenin, SOX2, MMP12, Vimentin, CD-44. Quantification of the transcripts’ level relative to the control by qPCR analyses. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). GAPDH considered as housekeeping gene. D RT-qPCR-verification of RNAseq data for differentially expressed genes in apoptotic pathway upon 50 nM TGS24 treatment in MDAMB-231 spheres for 24 h: NF-κβ, Sp1, GATA3, CASP9, PARP-1. Quantification of the transcripts’ level relative to the control by qPCR analyses. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). GAPDH considered as housekeeping gene. E Correlation of log2(fold-change) values from RNA-Seq and RT-PCR analysis. The R2 value is 0.854. F Interaction network of upregulated and downregulated genes, obtained using STRING database, with the minimum required interaction score of 0.400 and network edges representing evidence of an interaction
G4 formation at MAPK12 promoter thwarts oncogenic RAS transformation and inhibits the trans-activation of NANOG to reduce stemness in MDAMB-231 cells
While RNA-seq analyses provided a comprehensive overview of altered gene expression, the results may not solely attribute to MAPK12-G4 targeting as TGS24 or other NDI molecules bind to other parallel G4s. Therefore, further validation of the RNA-seq analysis is indeed required to confirm the role of MAPK12-G4 in stemness regulation. Breast cancer stem cells are characterized by high expression of CD44 markers over plasma membrane. Since MAPK12 downregulation by TGS24 treatment had been found to decrease tumorsphere formation in MDAMB-231 that potentially had a subpopulation of CSCs, flow cytometric analyses had been carried out to confirm if TGS24 treatment led to reduced stemness expansion in MDAMB-231 spheroids by the estimation of CD44High/CD24Low population. TGS24 treatment resulted into reduction of CD44High/CD24Low population by ~ 50% as compared to that of control. Consistent with NGS analyses, this observation confirmed that downregulation of G4-mediated MAPK12 expression by TGS24 led to stemness reduction in cancer cells (Fig. 6A). This finding was further supported by western blot study that revealed lower level of CD44, OCT-4, SOX2, and NANOG expression in MDAMB-231 spheres upon TGS24 treatment (Fig. 6H–J). On the other hand, TGS24 led to MAPK12-induced reduction in oncogenic RAS expression, which insinuated MAPK12-mediated positive feedback mechanism to regulate RAS expression (Fig. 6C). Previous studies showed that MAPK12 increases RAS expression through the activation of HSP90 (Heat shock protein 90), which forms a tetrameric complex of MAPK12, HSP90, RAS, and c-JUN and protects oncogenic RAS from proteasomal degradation, enhancing its stability [83, 84]. Therefore, in this study, HSP90 activation was closely inspected upon TGS24 treatment, to find out whether TGS24 reduced oncogenic RAS transformation by deactivating HSP90. Despite that HSP90 transcription remained unaffected upon TGS24 treatment, its protein expression was significantly reduced upon TGS24 treatment, accompanied with the reduction of RAS and c-JUN expression (Fig. 6C). Furthermore, the formation of tetrameric complex involving MAPK12, HSP90, RAS, and c-JUN was also investigated upon TGS24 treatment by co-immunoprecipitation studies, which revealed that TGS24 reduced RAS immunoprecipitation with MAPK12, HSP90, and c-JUN. Therefore, TGS24 reduced G4-mediated MAPK12 expression, which in turn downregulated RAS signaling in MDAMB-231 cells (through HSP90 deactivation) (Fig. 6D). Previous studies have shown that c‐JUN stimulated NANOG transcription through its binding to the AP‐1‐like site across the NANOG promoter, leading to the stemness expansion of colon cancer cells [85]. Similarly, MAPK12 is implicated in the trans-activation of MMP9 [72] and cyclin D1 [86], through c-JUN binding to their AP1-binding sites. Therefore, MAPK12 immunoprecipitants (c-JUN, HSP90, and RAS) were further analyzed for their complex formation at the NANOG promoter upon TGS24 treatment by chromatin immunoprecipitation studies (Fig. 6E–G). TGS24 abrogated the occupancy of MAPK12, RAS, HSP90, and c-JUN at the NANOG promoter, which envisaged that TGS24 reduced the trans-activation of NANOG expression by arresting MAPK12 expression and oncogenic RAS transformation (Fig. 6E–G). These observations collectively emphasized that the G4 motifs in MAPK12 promoter stringently regulate MAPK12 expression in cells that abrogates the crosstalk between oncogenic RAS transformation and the activation of stemness-related pathways.
Stabilization of MAPK12-G4 motifs by TGS24 enhances stemness-markers via HSP90-RAS-c-JUN-NANOG axis. A Flow cytometric analyses of CD44 and CD24 expression in MDAMB-231 spheres upon TGS24 (50 nM) treatment for 24 h. Box and whisker plot displays the percentage of CD44 low/CD24low and CD44high /CD24 low population upon TGS24 (50 nM) treatment for 24 h. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). B Violin plot showing MAPK12 expression in CD44high /CD24low population upon TGS24 (50 nM) treatment for 24 h. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). C Western blot analyses of the proteins in RAS pathway: RAS, c-JUN, HSP90, MAPK12 under TGS24 (50 nM) treatment for 24 h in MDAMB-231 spheres. GAPDH considered as housekeeping protein. D Co-immunoprecipitation analysis showing tetrameric complex of RAS, HSP90, MAPK12, and c-JUN. MDAMB-231 spheres that express RAS, are subjected to immunoprecipitation (IP) with anti-RAS antibody and to immunoblotting (IB) with anti-HSP90, anti-c-JUN, and anti-MAPK12 antibodies upon TGS24 (50 nM) treatment for 24 h. E ChIP analyses. ChIP results using anti-RAS, anti-HSP90, anti-c-JUN, and anti-MAPK12 antibodies suggesting MAPK12, c-JUN, RAS, and HSP90 enrichment at NANOG promoter under TGS24 (50 nM) treatment for 24 h in MDAMB-231 spheres. F Schematic representation of NANOG promoter region (AP1 site). Transcription initiation site is marked with an arrow. Position of the ChIP primers used in semi-quantitative and Real-time PCR reactions following immunoprecipitation is indicated. Input fraction indicates total DNA; negative control immunoprecipitation uses rabbit IgG showing no signal. G Quantification of MAPK12, RAS, c-JUN, and HSP90 binding at NANOG promoter by percentage of input method based on quantitative real-time PCR under aforementioned conditions. Error bars represent mean ± SE (N = 3). Statistical differences are determined by one-way ANOVA followed by Tukey–Kramer test (*P < 0.05, **P < 0.01, ***P < 0.001). H RT-PCR analyses. Expression profile of OCT-4, SOX2, and NANOG transcripts upon the treatment of TGS24 (50 nM) treatment for 24 h in MDAMB-231 spheres. I Quantification of the transcripts’ level of OCT-4, SOX2, and NANOG relative to the control by qPCR analyses. Error bars represent mean ± SE (N = 3). Statistical differences are determined compared to the control by two-tailed Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001). J Western blot analyses of OCT-4, SOX2, CD-44, and NANOG upon the treatment of TGS24 (50 nM) treatment for 24 h in MDAMB-231 spheres
Discussion
GC-rich regions act as the structural and functional ‘punctuation marks’ for gene-regulatory mechanisms. They are prone to develop secondary nucleic acid structures, such as G4s and i-motifs and interact with specific transcription factors to modulate gene transcription. MAPK12, which lately gained attention due to its aberrant expression in myriad of cancers, harbors repeats of G-rich elements within its promoter region that stringently regulates its transcription. In this study, two adjacently located G4 motifs were mapped upstream of the MAPK12 promoter region, separated by a spacer of 21 bp, of which GQ-1 exhibits higher propensity to fold into G4 structures, as compared to GQ-2. The presence of two tandem G4 structures imparted conformational dynamics within the MAPK12 promoter element, maintaining an equilibrium between duplex and G4 forms, regulated by Nucleolin and Sp1. Although Nucleolin induces G4 formation at MAPK12 promoter by arresting its promoter activation, Sp1 promotes MAPK12 transcription by inducing duplex formation. Their binding specificity at G4 elements appear to direct a homeostatic regulation of MAPK12 transcription. This was verified by CRISPR/Cas9 mediated disruption of neighboring G4 elements that resulted in undetectable MAPK12 level and cellular death. Since G4 structures have emerged as an attractive therapeutic target for its ability to modulate oncogene transcription, the G4 motifs at MAPK12 promoter provided a promising target to impede its expression in the cancer cells.
The design of MAPK12-specific inhibitors is highly challenging due to its structural similarities with other MAPK isoforms and is limited by promiscuous binding of MAPK12-targeting small molecules to MAPK14 and/or MAPK13. Hence, the design of small molecules that could restrict gene transcription by targeting the unique G4 structures at its promoter gained increasing acceptance. Considering the immense role of G4 structures to maintain a threshold level of MAPK12 transcripts within the cells, selective targeting of G4s by small molecules seems to be a promising avenue for therapeutic intervention. In this study, NDI derivatives were assessed for their promising anticancer properties and high-affinity binding to MAPK12-G4 topologies. However, fluorescence studies provided a read out to claim its slightly higher binding affinity to G4 structures at MAPK12 promoter compared to other G4s in the genome. Therefore, further elucidation of structure-based design and the understanding of structural dynamics in MAPK12 G4s are essential to minimize off-target interactions with other G4 structures within the cells. Mechanistic details of Nucleolin-induced G4 formation at MAPK12 promoter could provide further insights to design selective probes to target and arrest the conformational dynamics at MAPK12 promoter. In this connection, revelation of the guanine residues (G7, G10, G18, and G21) that mainly introduced dynamicity in the G4 structures were the important findings of this study. Nevertheless, the role of these guanine residues to mediate interaction with Sp1 and/or Nucleolin remains to be investigated further. In a recent study, MAPK12 has been reported to play significant role in hepatocyte regeneration after partial hepatectomy by inducing retinoblastoma phosphorylation during G0/G1 transition, thereby enabling the cells to escape from the quiescent stage in response to the stress stimuli [20]. This study envisaged the important role of MAPK12 to induce stemness properties in liver cancer cells. In our study, we investigated the role of these G4 structures at MAPK12 promoter to regulate stemness expansion in TNBC cells and xenograft models. To understand the molecular basis of G4-driven transcription inhibition and the downstream effects, we carried out complete transcriptome study that evidenced reduced expression of oncogenes associated with RAS, apoptotic pathway and stemness-related signaling. These findings revealed that G4-driven MAPK12 transcription not only regulates oncogenic RAS transformation, but also regulates the expression of multiple oncogenes leading to apoptosis regulation and stemness expansion of cancer cells. However, it is crucial to emphasize that we cannot definitively ascertain whether TGS24 exclusively interacts with the identified promoter G4s. This uncertainty underscores the intricate nature of G4 targeting and the need for further investigations. Our further cellular studies showed that MAPK12 transcription led to constitutive activation of RAS pathway by protecting and increasing the stability of RAS protein from proteasomal degradation. In this regard, MAPK12 formed a tetrameric protein complex with HSP90 and c-JUN, involving MAPK12, RAS, HSP90, and c-JUN to inhibit proteasomal degradation of RAS. G4-mediated silencing of MAPK12 transcription led to the deactivation of RAS pathway by disrupting the formation of tetrameric protein complex, leading to the proliferation arrest of the cancer cells. On the other hand, G4-driven attenuation of MAPK12 transcription strongly inhibited the binding occupancy of the tetrameric complex at NANOG promoter leading to the downregulation of NANOG trans-activation in cancer cells. Therefore, while our transcriptomic studies might be impacted by TGS24’s nonspecific binding to other G4 structures, our further cellular investigation aligning with the RNA-seq study reinforced the effect of MAPK12-G4 targeting in stemness regulation. Therefore, G4 structures might play an imperative role to place MAPK12 at the junction of RAS and NANOG-mediated signaling, wherein the expression level of MAPK12 transcripts determine the crosstalk between RAS and NANOG-pathways (Fig. 7).
Availability of data and materials
Please see the supporting information available online.
References
Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW et al (1994) A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372:739–746
Cuadrado A, Nebreda AR (2010) Mechanisms and functions of p38 MAPK signalling. Biochem J 429:403–417
Yin N, Qi X, Tsai S, Lu Y, Basir Z, Oshima K, Thomas JP, Myers CR, Stoner G, Chen G (2016) p38gamma MAPK is required for inflammation-associated colon tumorigenesis. Oncogene 35:1039–1048
Xu M, Wang S, Wang Y, Wu H, Frank JA, Zhang Z, Luo J (2018) Role of p38gamma MAPK in regulation of EMT and cancer stem cells. Biochim Biophys Acta Mol Basis Dis 1864:3605–3617
Fang Y, Wang J, Wang G, Zhou C, Wang P, Zhao S, Zhao S, Huang S, Su W, Jiang P et al (2017) Inactivation of p38 MAPK contributes to stem cell-like properties of non-small cell lung cancer. Oncotarget 8:26702–26717
Qi X, Yin N, Ma S, Lepp A, Tang J, Jing W, Johnson B, Dwinell MB, Chitambar CR, Chen G (2015) p38gamma MAPK is a therapeutic target for triple-negative breast cancer by stimulation of cancer stem-like cell expansion. Stem Cells 33:2738–2747
Sahu V, Mohan A, Dey S (2019) p38 MAP kinases: plausible diagnostic and prognostic serum protein marker of non small cell lung cancer. Exp Mol Pathol 107:118–123
Chen XF, Pan YS, Zheng B, Lu Q (2019) p38gamma overexpression promotes renal cell carcinoma cell growth, proliferation and migration. Biochem Biophys Res Commun 516:466–473
Wang F, Qi XM, Wertz R, Mortensen M, Hagen C, Evans J, Sheinin Y, James M, Liu P, Tsai S et al (2020) p38gamma MAPK is essential for aerobic glycolysis and pancreatic tumorigenesis. Cancer Res 80:3251
Su C, Sun Q, Liu S, Wang H, Feng L, Cao Y (2019) Targeting p38gamma to inhibit human colorectal cancer cell progression. Biochem Biophys Res Commun 517:172–179
Chen H, Wang X, Guo F, Li P, Peng D, He J (2019) Impact of p38gamma mitogen-activated protein kinase (MAPK) on MDA-MB-231 breast cancer cells using metabolomic approach. Int J Biochem Cell Biol 107:6–13
Zhang XH, Nam S, Wu J, Chen CH, Liu X, Li H, McKeithan T, Gong Q, Chan WC, Yin HH et al (2018) Multi-kinase inhibitor with anti-p38gamma activity in cutaneous T-Cell lymphoma. J Invest Dermatol 138:2377–2387
Sahu V, Nigam L, Agnihotri V, Gupta A, Shekhar S, Subbarao N, Bhaskar S, Dey S (2019) Diagnostic significance of p38 Isoforms (p38alpha, p38beta, p38gamma, p38delta) in head and neck squamous cell carcinoma: comparative serum level evaluation and design of novel peptide inhibitor targeting the same. Cancer Res Treat 51:313–325
Tang J, Qi X, Mercola D, Han J, Chen G (2005) Essential role of p38gamma in K-Ras transformation independent of phosphorylation. J Biol Chem 280:23910–23917
Hou SW, Zhi HY, Pohl N, Loesch M, Qi XM, Li RS, Basir Z, Chen G (2010) PTPH1 dephosphorylates and cooperates with p38gamma MAPK to increase ras oncogenesis through PDZ-mediated interaction. Cancer Res 70:2901–2910
Kwong J, Hong L, Liao R, Deng Q, Han J, Sun P (2009) p38alpha and p38gamma mediate oncogenic ras-induced senescence through differential mechanisms. J Biol Chem 284:11237–11246
Qi X, Pohl NM, Loesch M, Hou S, Li R, Qin JZ, Cuenda A, Chen G (2007) p38alpha antagonizes p38gamma activity through c-Jun-dependent ubiquitin-proteasome pathways in regulating Ras transformation and stress response. J Biol Chem 282:31398–31408
Qi X, Tang J, Loesch M, Pohl N, Alkan S, Chen G (2006) p38gamma mitogen-activated protein kinase integrates signaling crosstalk between Ras and estrogen receptor to increase breast cancer invasion. Cancer Res 66:7540–7547
Yin N, Lepp A, Ji Y, Mortensen M, Hou S, Qi XM, Myers CR, Chen G (2017) The K-Ras effector p38gamma MAPK confers intrinsic resistance to tyrosine kinase inhibitors by stimulating EGFR transcription and EGFR dephosphorylation. J Biol Chem 292:15070–15079
Tomas-Loba A, Manieri E, Gonzalez-Teran B, Mora A, Leiva-Vega L, Santamans AM, Romero-Becerra R, Rodriguez E, Pintor-Chocano A, Feixas F et al (2019) p38gamma is essential for cell cycle progression and liver tumorigenesis. Nature 568:557–560
Lovett FA, Cosgrove RA, Gonzalez I, Pell JM (2010) Essential role for p38alpha MAPK but not p38gamma MAPK in Igf2 expression and myoblast differentiation. Endocrinology 151:4368–4380
Sabio G, Arthur JS, Kuma Y, Peggie M, Carr J, Murray-Tait V, Centeno F, Goedert M, Morrice NA, Cuenda A (2005) p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J 24:1134–1145
Cuenda A, Sanz-Ezquerro JJ (2017) p38gamma and p38delta: from spectators to key physiological players. Trends Biochem Sci 42:431–442
Gaestel M, Kotlyarov A, Kracht M (2009) Targeting innate immunity protein kinase signalling in inflammation. Nat Rev Drug Discov 8:480–499
Yurtsever Z, Scheaffer SM, Romero AG, Holtzman MJ, Brett TJ (2015) The crystal structure of phosphorylated MAPK13 reveals common structural features and differences in p38 MAPK family activation. Acta Crystallogr D Biol Crystallogr 71:790–799
Kondoh Y, Honda K, Hiranuma S, Hayashi T, Shimizu T, Watanabe N, Osada H (2016) Comparative chemical array screening for p38gamma/delta MAPK inhibitors using a single gatekeeper residue difference between p38alpha/beta and p38gamma/delta. Sci Rep 6:29881
Eyers PA, Craxton M, Morrice N, Cohen P, Goedert M (1998) Conversion of SB 203580-insensitive MAP kinase family members to drug-sensitive forms by a single amino-acid substitution. Chem Biol 5:321–328
Lee MR, Dominguez C (2005) MAP kinase p38 inhibitors: clinical results and an intimate look at their interactions with p38alpha protein. Curr Med Chem 12:2979–2994
Zhang J, Shen B, Lin A (2007) Novel strategies for inhibition of the p38 MAPK pathway. Trends Pharmacol Sci 28:286–295
Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH (1997) Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem 40:2113–2116
Harrell WA Jr (2006) Quadruplex nucleic acids. The Royal Society of Chemistry, London
Hänsel-Hertsch R, Di Antonio M, Balasubramanian S (2017) DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol 18:279–284
Kumari S, Bugaut A, Huppert JL, Balasubramanian S (2007) An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol 3:218–221
Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP et al (2009) Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Can Res 69:7653–7661
Balasubramanian S, Hurley LH, Neidle S (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 10:261–275
Paeschke K, Capra JA, Zakian VA (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145:678–691
Lemmens B, van Schendel R, Tijsterman M (2015) Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat Commun 6:8909
Sengupta P, Bhattacharya A, Sa G, Das T, Chatterjee S (2019) Truncated G-Quadruplex isomers cross-talk with the transcription factors to maintain homeostatic equilibria in c-MYC transcription. Biochemistry 58:1975–1991
Ahn JK, Pitluk ZW, Ward DC (1992) The GC box and TATA transcription control elements in the P38 promoter of the minute virus of mice are necessary and sufficient for transactivation by the nonstructural protein NS1. J Virol 66:3776–3783
Ahmed AA, Marchetti C, Ohnmacht SA, Neidle S (2020) A G-quadruplex-binding compound shows potent activity in human gemcitabine-resistant pancreatic cancer cells. Sci Rep 10:12192
Olejko L, Bald I (2017) FRET efficiency and antenna effect in multi-color DNA origami-based light harvesting systems. RSC Adv 7:23924–23934
Fleming AM, Burrows CJ (2020) Interplay of guanine oxidation and G-quadruplex folding in gene promoters. J Am Chem Soc 142:1115–1136
Kikin O, D’Antonio L, Bagga PS (2006) QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 34:W676-682
Konig SL, Huppert JL, Sigel RK, Evans AC (2013) Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences. Nucleic Acids Res 41:7453–7461
Cogoi S, Shchekotikhin AE, Xodo LE (2014) HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res 42:8379–8388
Onel B, Carver M, Wu G, Timonina D, Kalarn S, Larriva M, Yang D (2016) A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J Am Chem Soc 138:2563–2570
Tucker BA, Hudson JS, Ding L, Lewis E, Sheardy RD, Kharlampieva E, Graves D (2018) Stability of the Na(+) form of the human telomeric g-quadruplex: role of adenines in stabilizing G-quadruplex structure. ACS Omega 3:844–855
Li YY, Macgregor RB Jr (2016) A thermodynamic study of adenine and thymine substitutions in the loops of the oligodeoxyribonucleotide HTel. J Phys Chem B 120:8830–8836
Guedin A, Alberti P, Mergny JL (2009) Stability of intramolecular quadruplexes: sequence effects in the central loop. Nucleic Acids Res 37:5559–5567
Gavathiotis E, Searle MS (2003) Structure of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat: evidence for A-tetrad formation from NMR and molecular dynamics simulations. Org Biomol Chem 1:1650–1656
Lightfoot HL, Hagen T, Tatum NJ, Hall J (2019) The diverse structural landscape of quadruplexes. FEBS Lett 593:2083–2102
Huppert JL (2008) Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem Soc Rev 37:1375–1384
Perez-Arnaiz C, Busto N, Santolaya J, Leal JM, Barone G, Garcia B (2018) Kinetic evidence for interaction of TMPyP4 with two different G-quadruplex conformations of human telomeric DNA. Biochim Biophys Acta Gen Subj 1862:522–531
Wei C, Jia G, Zhou J, Han G, Li C (2009) Evidence for the binding mode of porphyrins to G-quadruplex DNA. Phys Chem Chem Phys 11:4025–4032
Monchaud D, Granzhan A, Saettel N, Guedin A, Mergny JL, Teulade-Fichou MP (2010) “One ring to bind them all”-part I: the efficiency of the macrocyclic scaffold for g-quadruplex DNA recognition. J Nucleic Acids 2010:525862
Cheng MJ, Cao YG (2017) TMPYP4 exerted antitumor effects in human cervical cancer cells through activation of p38 mitogen-activated protein kinase. Biol Res 50:24
Morris MJ, Wingate KL, Silwal J, Leeper TC, Basu S (2012) The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res 40:4137–4145
Zhang YQ, Zhang YH, Xie J, Li MN, Liu ZR, Shen JY, Shi SS, Lan XY, Wang S, Cheng NL (2016) TMPyP4-regulated cell proliferation and apoptosis through the Wnt/beta-catenin signaling pathway in SW480 cells. J Recept Signal Transduct Res 36:167–172
Ohnmacht SA, Marchetti C, Gunaratnam M, Besser RJ, Haider SM, Di Vita G, Lowe HL, Mellinas-Gomez M, Diocou S, Robson M et al (2015) A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci Rep 5:11385
Marchetti C, Zyner KG, Ohnmacht SA, Robson M, Haider SM, Morton JP, Marsico G, Vo T, Laughlin-Toth S, Ahmed AA et al (2018) Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-Quadruplex-binding small molecule. J Med Chem 61:2500–2517
Micco M, Collie GW, Dale AG, Ohnmacht SA, Pazitna I, Gunaratnam M, Reszka AP, Neidle S (2013) Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J Med Chem 56:2959–2974
Pirota V, Nadai M, Doria F, Richter SN (2019) Naphthalene diimides as multimodal G-quadruplex-selective ligands. Molecules 24:426
Street STG, Chin DN, Hollingworth GJ, Berry M, Morales JC, Galan MC (2017) Divalent naphthalene diimide ligands display high selectivity for the human telomeric G-quadruplex in K(+) buffer. Chemistry 23:6953–6958
Basak S, Nandi N, Paul S, Banerjee A (2018) Luminescent naphthalene diimide-based peptide in aqueous medium and in solid state: rewritable fluorescent color code. ACS Omega 3:2174–2182
Vo T, Oxenford S, Angell R, Marchetti C, Ohnmacht SA, Wilson WD, Neidle S (2020) Substituted naphthalenediimide compounds bind selectively to two human quadruplex structures with parallel topology. ACS Med Chem Lett 11:991–999
McKnight RE, Reisenauer E, Pintado MV, Polasani SR, Dixon DW (2011) Substituent effect on the preferred DNA binding mode and affinity of a homologous series of naphthalene diimides. Bioorg Med Chem Lett 21:4288–4291
Peduto A, Pagano B, Petronzi C, Massa A, Esposito V, Virgilio A, Paduano F, Trapasso F, Fiorito F, Florio S et al (2011) Design, synthesis, biophysical and biological studies of trisubstituted naphthalimides as G-quadruplex ligands. Bioorg Med Chem 19:6419–6429
Van Quaquebeke E, Mahieu T, Dumont P, Dewelle J, Ribaucour F, Simon G, Sauvage S, Gaussin JF, Tuti J, El Yazidi M et al (2007) 2,2,2-Trichloro-N-({2-[2-(dimethylamino)ethyl]-1,3-dioxo-2,3-dihydro-1H-benzo[de] isoquinolin- 5-yl}carbamoyl)acetamide (UNBS3157), a novel nonhematotoxic naphthalimide derivative with potent antitumor activity. J Med Chem 50:4122–4134
Wang Y, Zhang X, Zhao J, Xie S, Wang C (2012) Nonhematotoxic naphthalene diimide modified by polyamine: synthesis and biological evaluation. J Med Chem 55:3502–3512
Huang J, Luo Q, Xiao Y, Li H, Kong L, Ren G (2017) The implication from RAS/RAF/ERK signaling pathway increased activation in epirubicin treated triple negative breast cancer. Oncotarget 8:108249–108260
Yu B, You W, Chen G, Yu Y, Yang Q (2019) MiR-140-5p inhibits cell proliferation and metastasis by regulating MUC1 via BCL2A1/MAPK pathway in triple negative breast cancer. Cell Cycle 18:2641–2650
Loesch M, Zhi HY, Hou SW, Qi XM, Li RS, Basir Z, Iftner T, Cuenda A, Chen G (2010) p38gamma MAPK cooperates with c-Jun in trans-activating matrix metalloproteinase 9. J Biol Chem 285:15149–15158
Long DL, Loeser RF (2010) p38gamma mitogen-activated protein kinase suppresses chondrocyte production of MMP-13 in response to catabolic stimulation. Osteoarthr Cartil 18:1203–1210
Sutherland C, Cui Y, Mao H, Hurley LH (2016) A mechanosensor mechanism controls the G-Quadruplex/i-Motif molecular switch in the MYC promoter NHE III1. J Am Chem Soc 138:14138–14151
Masuzawa T, Oyoshi T (2020) Roles of the RGG domain and RNA recognition motif of nucleolin in G-quadruplex stabilization. ACS Omega 5:5202–5208
Todd AK, Neidle S (2008) The relationship of potential G-quadruplex sequences in cis-upstream regions of the human genome to SP1-binding elements. Nucleic Acids Res 36:2700–2704
Lago S, Tosoni E, Nadai M, Palumbo M, Richter SN (2017) The cellular protein nucleolin preferentially binds long-looped G-quadruplex nucleic acids. Biochim Biophys Acta Gen Subj 1861:1371–1381
Sengar A, Vandana JJ, Chambers VS, Di Antonio M, Winnerdy FR, Balasubramanian S, Phan AT (2019) Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter. Nucleic Acids Res 47:1564–1572
Qiu J, Liu J, Chen S, Ou T-M, Tan J-H, Gu L-Q, Huang Z-S, Li D (2015) Role of Hairpin-Quadruplex DNA Secondary Structural Conversion in the Promoter of hnRNP K in Gene Transcriptional Regulation. Org Lett 17:4584–4587
Lyu J, Shao R, Yung PYK, Elsässer SJ (2022) Genome-wide mapping of G-quadruplex structures with CUT & Tag. Nucleic Acids Res 50:e13
Esnault C, Magat T, Zine El Aabidine A et al (2023) G4access identifies G-quadruplexes and their associations with open chromatin and imprinting control regions. Nat Genet 55:1359–1369
Ahmed AA, Angell R, Oxenford S, Worthington J, Williams N, Barton N, Fowler TG, O’Flynn DE, Sunose M, McConville M et al (2020) Asymmetrically substituted quadruplex-binding naphthalene diimide showing potent activity in pancreatic cancer models. ACS Med Chem Lett 11:1634–1644
Qi X, Xie C, Hou S, Li G, Yin N, Dong L, Lepp A, Chesnik MA, Mirza SP, Szabo A et al (2014) Identification of a ternary protein-complex as a therapeutic target for K-Ras-dependent colon cancer. Oncotarget 5:4269–4282
Trepel J, Mollapour M, Giaccone G, Neckers L (2010) Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer 10:537–549
Ibrahim EE, Babaei-Jadidi R, Saadeddin A, Spencer-Dene B, Hossaini S, Abuzinadah M, Li N, Fadhil W, Ilyas M, Bonnet D et al (2012) Embryonic NANOG activity defines colorectal cancer stem cells and modulates through AP1- and TCF-dependent mechanisms. Stem Cells 30:2076–2087
Qi X, Zhi H, Lepp A, Wang P, Huang J, Basir Z, Chitambar CR, Myers CR, Chen G (2012) p38gamma mitogen-activated protein kinase (MAPK) confers breast cancer hormone sensitivity by switching estrogen receptor (ER) signaling from classical to nonclassical pathway via stimulating ER phosphorylation and c-Jun transcription. J Biol Chem 287:14681–14691
Acknowledgements
We thank the Central Instrumental Facility, Bose Institute. We acknowledge the Medgenome company for cultivating NGS data. We thank CSIR and Bose Institute, Department of Science and Technology, Government of India for student’s fellowship. We also thank DBT, Govt of India for funding support. We thank Mr. Hariharan Moorthy (Bioorganic Chemistry Laboratory, New Chemistry Unit, Jawaharlal Nehru Centre for Advanced Scientific Research) for UV spectroscopy experimental support.
Funding
SC thanks the DBT, New Delhi, Government of India BT/PR28663/MED/30/2020/2018 for research grant. TG thanks JNCASR (TRC II: JNC/2022/01653) and SERB, New Delhi, Government of India (CRG/2020/004594) for research grants.
Author information
Authors and Affiliations
Contributions
PS, AD and YVS performed the experiments and wrote the manuscript. AD, AD, SH and KJ worked in animal models. NB, AD, TRC performed experiments. PS, AD, SC and TG conceived the idea. PS, YVS, SC, NB, and TG analysed the data. PS, AD, TG, SC wrote the paper.
Corresponding authors
Ethics declarations
Conflict of Interest
Authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sengupta, P., Dutta, A., Suseela, Y.V. et al. G-quadruplex structural dynamics at MAPK12 promoter dictates transcriptional switch to determine stemness in breast cancer. Cell. Mol. Life Sci. 81, 33 (2024). https://doi.org/10.1007/s00018-023-05046-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-05046-6