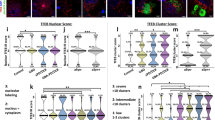
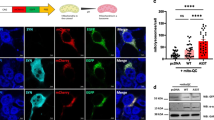
Abstract
Huntingtin-associated protein 1 (HAP1) is the first identified protein whose function is affected by its abnormal interaction with mutant huntingtin (mHTT), which causes Huntington disease. However, the expression patterns of Hap1 and Htt in the rodent brain are not correlated. Here we found that the primate HAP1, unlike the rodent Hap1, is correlatively expressed with HTT in the primate brains. CRISPR/Cas9 targeting revealed that HAP1 deficiency in the developing human neurons did not affect neuronal differentiation and gene expression as seen in the mouse neurons. However, deletion of HAP1 exacerbated neurotoxicity of mutant HTT in the organotypic brain slices of adult monkeys. These findings demonstrate differential HAP1 expression and function in the mouse and primate brains, and suggest that interaction of HAP1 with mutant HTT may be involved in mutant HTT-mediated neurotoxicity in adult primate neurons.









Similar content being viewed by others
Data availability
The raw data from this publication have been deposited. The RNA sequencing data that support the findings of this study are available in NCBI’s sequence read archive (SRA) database through Bioproject Accession number: PRJNA796166 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA796166/).
References
Li XJ, Li SH, Sharp AH et al (1995) A huntingtin-associated protein enriched in brain with implications for pathology. Nature 378:398–402. https://doi.org/10.1038/378398a0
Gauthier LR, Charrin BC, Borrell-Pagès M et al (2004) Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118:127–138. https://doi.org/10.1016/j.cell.2004.06.018
Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL et al (2010) Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron 65:53–65. https://doi.org/10.1016/j.neuron.2009.12.007
Keryer G, Pineda JR, Liot G et al (2011) Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest 121:4372–4382. https://doi.org/10.1172/jci57552
Roux JC, Zala D, Panayotis N et al (2012) Modification of Mecp2 dosage alters axonal transport through the Huntingtin/Hap1 pathway. Neurobiol Dis 45:786–795. https://doi.org/10.1016/j.nbd.2011.11.002
Wong YC, Holzbaur EL (2014) The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci 34:1293–1305. https://doi.org/10.1523/jneurosci.1870-13.2014
Mackenzie KD, Duffield MD, Peiris H et al (2014) Huntingtin-associated protein 1 regulates exocytosis, vesicle docking, readily releasable pool size and fusion pore stability in mouse chromaffin cells. J Physiol 592:1505–1518. https://doi.org/10.1113/jphysiol.2013.268342
Fujinaga R, Kawano J, Matsuzaki Y et al (2004) Neuroanatomical distribution of Huntingtin-associated protein 1-mRNA in the male mouse brain. J Comp Neurol 478:88–109. https://doi.org/10.1002/cne.20277
Hedreen JC, Folstein SE (1995) Early loss of neostriatal striosome neurons in Huntington’s disease. J Neuropathol Exp Neurol 54:105–120. https://doi.org/10.1097/00005072-199501000-00013
Dom R, Malfroid M, Baro F (1976) Neuropathology of Huntington’s chorea. studies of the ventrobasal complex of the thalamus. Neurology 26:64–68. https://doi.org/10.1212/wnl.26.1.64
Murphy KP, Carter RJ, Lione LA et al (2000) Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington’s disease mutation. J Neurosci 20:5115–5123. https://doi.org/10.1523/jneurosci.20-13-05115.2000
Nasir J, Floresco SB, O’Kusky JR et al (1995) Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell 81:811–823. https://doi.org/10.1016/0092-8674(95)90542-1
Duyao MP, Auerbach AB, Ryan A et al (1995) Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science 269:407–410. https://doi.org/10.1126/science.7618107
Zeitlin S, Liu JP, Chapman DL et al (1995) Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet 11:155–163. https://doi.org/10.1038/ng1095-155
Chan EY, Nasir J, Gutekunst CA et al (2002) Targeted disruption of Huntingtin-associated protein-1 (Hap1) results in postnatal death due to depressed feeding behavior. Hum Mol Genet 11:945–959. https://doi.org/10.1093/hmg/11.8.945
Li SH, Yu ZX, Li CL et al (2003) Lack of huntingtin-associated protein-1 causes neuronal death resembling hypothalamic degeneration in Huntington’s disease. J Neurosci 23:6956–6964. https://doi.org/10.1523/jneurosci.23-17-06956.2003
Xiang J, Yang H, Zhao T et al (2014) Huntingtin-associated protein 1 regulates postnatal neurogenesis and neurotrophin receptor sorting. J Clin Invest 124:85–98. https://doi.org/10.1172/jci69206
Sheng G, Chang GQ, Lin JY et al (2006) Hypothalamic huntingtin-associated protein 1 as a mediator of feeding behavior. Nat Med 12:526–533. https://doi.org/10.1038/nm1382
Xiang J, Yan S, Li SH et al (2015) Postnatal loss of hap1 reduces hippocampal neurogenesis and causes adult depressive-like behavior in mice. PLoS Genet 11:e1005175. https://doi.org/10.1371/journal.pgen.1005175
Li XJ, Sharp AH, Li SH et al (1996) Huntingtin-associated protein (HAP1): discrete neuronal localizations in the brain resemble those of neuronal nitric oxide synthase. Proc Natl Acad Sci USA 93:4839–4844. https://doi.org/10.1073/pnas.93.10.4839
Li SH, Hosseini SH, Gutekunst CA et al (1998) A human HAP1 homologue. Cloning, expression, and interaction with huntingtin. J Biol Chem 273:19220–19227. https://doi.org/10.1074/jbc.273.30.19220
Yang S, Chang R, Yang H et al (2017) CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J Clin Invest 127:2719–2724. https://doi.org/10.1172/jci92087
Li T, Li S, Gao X et al (2019) Expression and localization of Huntingtin-Associated Protein 1 (HAP1) in the human digestive system. Dig Dis Sci 64:1486–1492. https://doi.org/10.1007/s10620-018-5425-5
Evers MM, Schut MH, Pepers BA et al (2015) Making (anti-) sense out of huntingtin levels in Huntington disease. Mol Neurodegener 10:21. https://doi.org/10.1186/s13024-015-0018-7
Vigont VA, Grekhnev DA, Lebedeva OS et al (2021) STIM2 mediates excessive store-operated calcium entry in patient-specific iPSC-derived neurons modeling a juvenile form of Huntington’s disease. Front Cell Dev Biol 9:625231. https://doi.org/10.3389/fcell.2021.625231
Li SH, Li H, Torre ER et al (2000) Expression of huntingtin-associated protein-1 in neuronal cells implicates a role in neuritic growth. Mol Cell Neurosci 16:168–183. https://doi.org/10.1006/mcne.2000.0858
Gutekunst CA, Li SH, Yi H et al (1998) The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): comparison with huntingtin in rat and human. J Neurosci 18:7674–7686. https://doi.org/10.1523/jneurosci.18-19-07674.1998
Li SH, Gutekunst CA, Hersch SM et al (1998) Association of HAP1 isoforms with a unique cytoplasmic structure. J Neurochem 71:2178–2185. https://doi.org/10.1046/j.1471-4159.1998.71052178.x
Nguyen GD, Gokhan S, Molero AE et al (2013) Selective roles of normal and mutant huntingtin in neural induction and early neurogenesis. PLoS ONE 8:e64368. https://doi.org/10.1371/journal.pone.0064368
Chen X, Xin N, Pan Y et al (2020) Huntingtin-associated protein 1 in mouse hypothalamus stabilizes glucocorticoid receptor in stress response. Front Cell Neurosci 14:125. https://doi.org/10.3389/fncel.2020.00125
Liu Q, Cheng S, Yang H et al (2020) Loss of Hap1 selectively promotes striatal degeneration in Huntington disease mice. Proc Natl Acad USA 117:20265–20273. https://doi.org/10.1073/pnas.2002283117
Scholzen T, Gerdes J (2000) The Ki-67 protein: from the known and the unknown. J Cell Physiol 182:311–322. https://doi.org/10.1002/(sici)1097-4652(200003)182:3%3c311::aid-jcp1%3e3.0.co;2-9
Zhao T, Hong Y, Li S et al (2016) Compartment-dependent degradation of mutant huntingtin accounts for its preferential accumulation in neuronal processes. J Neurosci 36:8317–8328. https://doi.org/10.1523/JNEUROSCI.0806-16.2016
Sun YM, Zhang YB, Wu ZY (2017) Huntington’s disease: relationship between phenotype and genotype. Mol Neurobiol 54:342–348. https://doi.org/10.1007/s12035-015-9662-8
Bhide PG, Day M, Sapp E et al (1996) Expression of normal and mutant huntingtin in the developing brain. J Neurosci 16:5523–5535. https://doi.org/10.1523/jneurosci.16-17-05523.1996
Li SH, Schilling G, Young WS 3rd et al (1993) Huntington’s disease gene (IT15) is widely expressed in human and rat tissues. Neuron 11:985–993. https://doi.org/10.1016/0896-6273(93)90127-d
Harjes P, Wanker EE (2003) The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci 28:425–433. https://doi.org/10.1016/s0968-0004(03)00168-3
Li SH, Li XJ (2004) Huntingtin-protein interactions and the pathogenesis of Huntington’s disease. Trends Genet 20:146–154. https://doi.org/10.1016/j.tig.2004.01.008
Borrell-Pagès M, Zala D, Humbert S et al (2006) Huntington’s disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci 63:2642–2660. https://doi.org/10.1007/s00018-006-6242-0
Wroblewski G, Islam MN, Yanai A et al (2018) Distribution of HAP1-immunoreactive cells in the retrosplenial-retrohippocampal area of adult rat brain and its application to a refined neuroanatomical understanding of the region. Neuroscience 394:109–126. https://doi.org/10.1016/j.neuroscience.2018.10.029
Reiner A, Dragatsis I, Zeitlin S et al (2003) Wild-type huntingtin plays a role in brain development and neuronal survival. Mol Neurobiol 28:259–276. https://doi.org/10.1385/mn:28:3:259
Caviston JP, Holzbaur EL (2009) Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol 19:147–155. https://doi.org/10.1016/j.tcb.2009.01.005
Rong J, Li SH, Li XJ (2007) Regulation of intracellular HAP1 trafficking. J Neurosci Res 85:3025–3029. https://doi.org/10.1002/jnr.21326
Yin P, Guo X, Yang W et al (2019) Caspase-4 mediates cytoplasmic accumulation of TDP-43 in the primate brains. Acta Neuropathol 137:919–937. https://doi.org/10.1007/s00401-019-01979-0
Yang W, Guo X, Tu Z et al (2022) PINK1 kinase dysfunction triggers neurodegeneration in the primate brain without impacting mitochondrial homeostasis. Protein Cell 13:26–46. https://doi.org/10.1007/s13238-021-00888-x
Yan XX, Ma C, Bao AM et al (2015) Brain banking as a cornerstone of neuroscience in China. Lancet Neurol 14:136. https://doi.org/10.1016/s1474-4422(14)70259-5
Qiu W, Zhang H, Bao A et al (2019) Standardized operational protocol for human brain banking in China. Neurosci Bull 35:270–276. https://doi.org/10.1007/s12264-018-0306-7
Zhang XY, Li J, Li CJ et al (2021) Differential development and electrophysiological activity in cultured cortical neurons from the mouse and cynomolgus monkey. Neural Regen Res 16:2446–2452. https://doi.org/10.4103/1673-5374.313056
Patro R, Duggal G, Love MI et al (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. https://doi.org/10.1038/nmeth.4197
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. https://doi.org/10.1093/nar/gks042
Chen Y, Lun AT, Smyth GK (2016) From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 5:1438. https://doi.org/10.12688/f1000research.8987.2
Yu G, Wang LG, Han Y et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Acknowledgements
We thank Professor Xiao-Xin Yan at Xiangya School of Medicine, Central South University, Changsha, China for providing human postmortem brain tissues, Yuefeng Li at Guangdong Landao Biotechnology Co. Ltd for animal care and experiment. This work was supported by The National Natural Science Foundation of China (81830032, 82071421, 31872779, 81901289), Guangzhou Key Research Program on Brain Science (202007030008), Department of Science and Technology of Guangdong Province (2021ZT09Y007; 2020B121201006; 2018B030337001). XC was funded by Science and Technology Research Project of Education Department of Hubei Province (B2020020). This study was supported by the high performance public computing service platform of Jinan University.
Funding
This research was funded by National Natural Science Foundation of China, Grant numbers [82071421, 81830032, 31872779].
Author information
Authors and Affiliations
Contributions
SL and X-JL designed experiments. XC performed all the experiments with the help of YS. LC performed RNA-seq data analysis. X-SC conducted monkey cortical neurons culture. MP conducted organotypic monkey brain slice culture. YZ and QW aided in molecular biological experiments. WY, PY, DH, XG, SY, YZ and S Yan provided important advice, reagents, and experimental samples. XC, X-J L and SL wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, X., Sun, Y., Chen, L. et al. Differential expression and roles of Huntingtin and Huntingtin-associated protein 1 in the mouse and primate brains. Cell. Mol. Life Sci. 79, 554 (2022). https://doi.org/10.1007/s00018-022-04577-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04577-8