Abstract
The dynamic transition between epithelial-like and mesenchymal-like cell states has been a focus for extensive investigation for decades, reflective of the importance of Epithelial-Mesenchymal Transition (EMT) through development, in the adult, and the contributing role EMT has to pathologies including metastasis and fibrosis. Not surprisingly, regulation of the complex genetic networks that underlie EMT have been attributed to multiple transcription factors and microRNAs. What is surprising, however, are the sheer number of different regulators (hundreds of transcription factors and microRNAs) for which critical roles have been described. This review seeks not to collate these studies, but to provide a perspective on the fundamental question of whether it is really feasible that so many regulators play important roles and if so, what does this tell us about EMT and more generally, the genetic machinery that controls complex biological processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epithelial-Mesenchymal Transition (EMT) describes a process by which epithelial cells, possessing apical-basal polarity and characterized by stable cell–cell and cell-basement interactions, acquire mesenchymal characteristics including a fibroblast-like morphology, a stress-fibre cytoarchitecture and increased migratory capacity [1,2,3,4]. First described during early embryogenesis [5], it is now widely recognised that EMT, and the reverse process of mesenchymal to epithelial transition (MET), occur widely not only throughout development [6,7,8] but also in the adult, facilitating key processes such as wound healing [9] and driving pathologies including fibrosis [10,11,12] and cancer metastasis when inappropriately regulated [3, 13,14,15,16].
EMT, or Epithelial-Mesenchymal Plasticity (EMP) as it is also called to reflect its reversible and dynamic nature, is often described as being regulated via a small number of core transcription factors (TF) and by extension, a select group of microRNAs (miRNAs) with which these TFs participate in regulatory feedback interactions [17, 18]. Across the literature, however, hundreds of different TFs and miRNAs have been individually implicated as driving EMT/MET. That is to say, the individual manipulation of hundreds of separate TFs or miRNAs result in a reported EMT or MET phenotypic change as indicated by cell morphology, the altered expression of EMT marker genes (such as the E-cadherin to N-cadherin switch) and changes to the migratory and invasive capacity of cells.
In this review, we seek not to simply catalog these studies (indeed their vast number would make such a task impractical), but to ask a more fundamental question of is it really feasible there exist so many regulators of a biological process such as EMT? If many hundreds of direct regulators seem implausible, why have so many been implicated? Alternately, if hundreds of regulators are a biological reality, why is such complexity required and what challenges might this pose for attempts to manipulate EMT as a therapeutic strategy?
To EMT or not to EMT: a question of definition?
One of the confounding factors that likely contributes to the vast array of reported EMT regulators lies with the very definition of EMT itself. For example, is it sufficient to ascribe EMT/MET based upon key marker gene expression alone or are phenotypic changes also required? If so, what genes and what effects constitute a minimum threshold? This question is made all the more difficult by the growing realization that EMT is not a separation between two alternate states, but rather a continuum of partial or hybrid EMT states with the intermediate nature of the state itself central to the phenotype [19,20,21,22,23,24].
A hallmark of EMT are morphological and cytoskeletal changes that alter cell–cell and cell–matrix contacts. These include the downregulation of key genes such as CDH1 (E-cadherin) and CRB3 (Crumbs3), contributing to the loss of adherens and tight junctions, respectively [15]. The loss of E-cadherin is often accompanied by “cadherin-switching” [25], facilitating motility through the upregulation of N-cadherin which mediates more flexible cell–cell contacts. EMT further enhances motility by the cytoskeletal rearrangements that promote focal adhesions (crosslinking actin filaments to integrins) and invadopodia (where matrix metalloproteases degrade an extracellular matrix otherwise unconducive to motility) [15]. These events are thought to underlie metastasis, with which EMT has been extensively linked. Indeed, tumour cells at both the invasive front [26,27,28,29,30,31] and within the circulation [32,33,34,35,36,37] often lose epithelial and/or gain mesenchymal markers and the expression of EMT-promoting TFs often correlates with poor clinical outcome [38, 39] and drives metastasis in animal models [40,41,42,43]. It may seem reasonable to require such evidence to be presented when claiming an EMT-regulatory role for a new gene or stimulus, however, the causative link between EMT and metastasis remains controversial [16], the morphological and cytoskeletal appearance of cells undergoing EMT varies widely and the inference that changes in the (often two-dimensional) motility of cells in vitro reflects events in vivo is problematic [44]. Further, the different phenotypic outcomes of EMT have been expanded beyond the traditionally reported effects on morphology and motility and into areas including stemness, chemoresistance and immunosuppression [2, 13]. Outcomes, however, are not universal, with specific phenotypes being more or less prominent depending upon the context.
Consistent with the breadth of EMT-associated phenotypes is the magnitude of the underlying transcriptional changes. Typically, the expression of thousands of genes is altered between epithelial and mesenchymal states, and though efforts have been made to deduce a core EMT signature [45,46,47,48,49,50], even among very well-established drivers of the process there is significant variation between different models. Another inherent difficulty relying upon marker genes is the assumption that changes in the expression of a subset of genes are providing a readout of a wider EMT process. This is especially problematic given the multiple processes that are associated with EMT and the prevalence of hybrid states.
These issues have led to the recent publication of a consensus statement on behalf of the EMT International Association (TEMTIA) [4], which aims to improve guidelines and definitions for EMT researchers in which it was recommended a combination of molecular markers and cellular changes should be required to define EMT. The nature of the markers and cellular changes required to demonstrate EMT/MET, however, are impossible to codify given the broad spectra of phenotypic outcomes and the inherent variation between cells in the genes that drive these changes. As such, defining rigid minimum criteria that one must meet to demonstrate EMT would seem impossible which in turn leaves open the door to claims of genes being “EMT regulators” when they actually regulate narrow aspects of EMT or regulate largely EMT-independent processes that nevertheless overlap or fall within the wider EMT realm. How many aspects of EMT must a gene regulate to be classed as an “EMT regulator” is, therefore, an open question.
Experimental design must also be taken into account when assessing the quality of any given study, especially if that study relies upon single or poorly controlled siRNAs or supraphysiological levels of expression. Even with these caveats, however, it remains true that for hundreds of regulatory TFs, miRNAs and lncRNAs, claims are made of their regulation of EMT, citing as evidence both marker genes and phenotypic changes and employing both exogenous expression and endogenous inhibition to do so. We contend therefore that questions of definition or quality of study are insufficient to dismiss the bulk of EMT/MET regulators that are reported, which in turn posits the question, why are there so many regulators of EMT?
EMT: interconnected layers of complexity
EMT inducing stimuli
EMT is induced when epithelial cells encounter specific signals, the best studied of which being the TGFβ proteins (TGFβ1,2,3)—a subset of a wider TGFβ superfamily that also includes bone morphogenic proteins (BMPs), growth differentiation factors (GDFs), activins and inhibins [51]. Many of these promote EMT in various developmental contexts including mesoderm formation [52], heart development [53, 54], neural crest delamination [55] and palate fusion [56]. Both the TGFβs and BMPs also promote fibrosis within the lung [57], liver [58] and kidney [59] and have been widely associated with enhancing plasticity and invasiveness during cancer dissemination [15]. Whilst being the most extensively studied, TGFβ is but one of the dozens of EMT-inducing stimuli, including other growth factors, cytokines and ligands that initiate signaling events through the binding and activation of cell-surface receptors. Prominent examples include the epidermal growth factor (EGF) [60], hepatocyte growth factor (HGF) [61], fibroblast growth factor (FGF) [62], vascular endothelial growth factor (VEGF) [63], insulin-like growth factor (IGF) [64], inflammatory mediators such as IL-8 [65] and ligands activating Notch [8, 66], Hedgehog [67] and Wnt [68, 69] signaling pathways. Additionally, EMT can be stimulated via non-growth-factor stimuli including hypoxia [70, 71], mechanical stress [72] and the metabolite oxalate [73].
Core EMT-regulating TFs
The large number of EMT-inducing stimuli initiate gene expression programs that involve and are driven by a broad array of TFs. Direct repression of the CDH1 gene (encoding E-cadherin) by SNAI1 was initially identified as a mechanism to drive EMT [74, 75]. Additional TFs have since been identified that also promote EMT, at least in part through direct CDH1 repression. These include the Snail family member SNAI2 [76], the ZEB family TFs ZEB1 and ZEB2 [77, 78] and a host of additional TFs including TBXT [79], E47 [80] and KLF8 [81]. EMT promoting TFs that work via mechanisms independent of direct CDH1 transcriptional repression are also established with better characterized examples including TWIST1 and TWIST2 [82], PRRX1 [83], GSC [84], TCF4 [85], SIX1 [86], FOXC2 [87] and SOX4 [88]. These differing mechanisms result in differing properties. The SNAIL and ZEB TFs for example are potent suppressors of the epithelial phenotype (consistent with direct suppression of CDH1), whilst TWIST and PRRX1 are more potent mesenchymal inducers [2]. Working in opposition are other TFs that enforce an epithelial phenotype including OVOL1 and OVOL2 [89], GRHL2 [90], p53 [91], ELF5 [92], FOXO3 [93] and FOXA1 [94].
The listed TFs, however, only represent a small number of those that have been directly implicated as driving either EMT or MET. Some of these are only mentioned sporadically whilst others are referenced in the majority of EMT studies. Of the best characterized TFs, it is the Snail, Zeb and Twist families that have become recognized as “core” EMT drivers which orchestrate widespread gene expression responses, including supporting the expression of each other. For example, TGFβ promotes the rapid upregulation of SNAI1 and SNAI2 in a manner dependent upon the SMADs and HMGA2 [95, 96] which in turn upregulates ZEB, the expression of which can then be maintained by autocrine TGFβ production which promotes mesenchymal stability [97]. Similarly, both SNAI1 and TWIST1 co-operate in the regulation of ZEB1 to promote EMT [98]. There are, however, many instances of non-redundant functions where the suppression of a single EMT-promoting TF is sufficient to block or severely curtail EMT and metastasis in experimental models without compensation by other core TFs [40, 41, 99,100,101,102]. There are also suggestions that specific sub-roles exist between core EMT TFs. For example, SNAI1 and/or SNAI2 are specifically associated with the resistance to chemotherapy [103, 104] whilst ZEB1 prevents apoptotic cells death [105,106,107]. There are even examples of family members playing opposing roles depending upon context [108]. For example, ZEB1 promotes the initiation and metastatic progression of melanoma which is supported by TWIST1, whilst ZEB2, supported by SNAI2, acts as a melanoma tumour suppressor [109]. The ZEBs also have opposing roles in osteoblast growth and differentiation [110] whilst SNAI1 and SNAI2 differentially regulate stemness and oncogenesis in cells of mammary and thyroid origin [111].
Thus, even at the most basic level of gene regulation in EMT—the actions of a handful of core mesenchymal-promoting TFs, complexities underlie the differential functions of these related family members. The explanations for this are various. For example, the unique targeting of genes resulting from small differences in their “E-box” DNA recognition motifs or the requirement for single or paired E-boxes at varied spacing on account of the positioning of the single (SNAIL) or paired (ZEB, TWIST) DNA-binding zinc fingers. Their regulation of genes is also influenced by their capacity to act as either transcriptional repressors or activators, which can alternate depending upon the cohort of cofactors with which they interact [112]. One such example was recently demonstrated with ZEB1. Although best characterized as a transcriptional repressor via direct binding to DNA at E-box motifs, ZEB1 can be recruited to a co-activator complex through interaction with the AP1 factors FOSL1 and JUN and the Hippo pathway TF, YAP1 [113]. Both actions of ZEB1 functionally synergise as ZEB represses epithelial genes and tumour suppressors and promotes the expression of oncogenes and EMT inducers including TGFβ1 and PTPN14, both genes that encode proteins capable of initiating EMT in their own right [114, 115].
Co-regulatory relationships between TFs and miRNAs
From the early days of miRNA network biology, it was reported that TFs were enriched among miRNA-predicted targets [116, 117] and TFs frequently form “hub” or key nodes within miRNA regulatory networks [118,119,120]. Such networks include both feedforward loops, whereby either the TF or miRNA regulate the other whilst both regulate a common downstream target, and feedback inhibition where the TF and miRNA both directly suppress the expression of the other at the transcriptional and post-transcriptional levels, respectively [121].
Gene circuits of this nature are widespread beyond EMT as they may reduce signaling noise [122, 123] and establish mutually exclusive phenotypic states; the most obvious example represented by the phenotypic balance that exists between the epithelially expressed miR-200 family of miRNAs and the mesenchymal promoting ZEB TFs which exist in a direct negative feedback relationship [124, 125]. Similar well-established feedback mechanisms exist between SNAI1 and miR-34 [126], SNAI1 and miR-203 [127] and SNAI2 and miR-200 [128]. Additional, more complex gene regulatory networks (GRNs) also exist. For example, the sequentially expressed EMT-promoting TFs SNAI1 and PRRX1, negatively regulate each other via a feedback loop involving miR-15, where SNAI1 directly represses PRRX1 transcription, whilst PRRX1 transcriptionally activates miR-15, a SNAI1-targeting miRNA [129]. Feedback motifs such as these exist within complex webs of TF:TF and TF:miRNA interactions. One study to illustrate this found that siRNAs targeted against 117 different TFs blocked TGF-β induced EMT in NMuMg cells (as determined by high throughput microscopy assessing cytoskeletal hallmarks of EMT—actin stress fibres, focal adhesions and fibronectin patches) [130]. Coupled with a similar (though more limited) screen for miRNAs that influence EMT in the same model system [131], a connected network of 46 TFs and 13 miRNAs were suggested to regulate EMT, each linked within a web of predicted positive and negative feedback loops that included 4 particularly important TF signaling hubs (ZEB1, TEAD2, FOSL2 and SOX4).
One of the key features of miRNAs is their capacity to simultaneously regulate large cohorts of genes, afforded by the short, and therefore frequently occurring, length of sequence complementarity through which they interact with their targets [132]. For example, miR-200 targets networks of genes associated with the dynamic regulation of the cytoskeleton which is a key component of EMT [133,134,135] and targets networks of genes downstream of the TGFβ and EGF receptors, perhaps the two best established EMT-inducing stimuli [136]. When coupled with the direct regulation of key transcriptional regulators it is through this “two-punch” mechanism (direct regulation of both the transcriptional regulators and the downstream non-TF network components) that miRNAs exert profound regulatory effects on gene expression. It is worth noting the extent to which the influence of miRNAs is mediated not just directly through their primary target genes, but also indirectly via the targets of the TFs that the miRNAs directly regulate. For example, examining the profile of Dicer-knockout fibroblasts (in which miRNAs are globally depleted), revealed a predominant effect on gene expression at the transcriptional level, both in terms of the number and degree of gene expression changes [137]. This has also been demonstrated specifically with regard to EMT, where both the expression or inhibition of miR-200 resulted in a series of co-ordinated transcriptional responses that were central to MET/EMT and that were likely the result of not only the direct regulation of ZEB but other TFs as well [136].
Alternative splicing
Although not the primary focus of this review, it is worth noting that the complex networks of TFs and miRNAs that regulate both each other and various EMT-associated genes are themselves embedded within additional mechanistic levels of regulation comprising alternative splicing, translational regulation and post-translational protein modification that affects protein stability and subcellular localization (Fig. 1).
Epithelial and mesenchymal cells show distinct alternative splicing patterns [138], regulated by splicing factors whose expression are controlled by EMT-TFs and miRNAs. An epithelial splicing pattern is primarily enforced by two paralogous RNA binding proteins, ESRP1 (epithelial-splicing regulatory protein 1) and ESRP2 which recognize a core UGG motif to guide exon skipping or inclusion depending upon whether the binding site is up- or downstream of the splicing junction [139,140,141]. In so doing, the ESRPs directly drive several hundred epithelial-specific splicing events, including the production of a shorter isoform of CTNND1 (p120 catenin) that promotes an epithelial phenotype by stabilizing E-cadherin at the cell membrane [142]. During EMT, the ESRP genes are directly downregulated by SNAI1, ZEB1 and ZEB2 [143, 144] whilst other RNA-binding proteins such as QKI (quaking), MBNL1 (muscleblind-like splicing regulator 1) and RBFOX (RNA-binding Fox-1 homolog) promote mesenchymal-specific splicing events [138, 139, 145,146,147] and guide circular RNA formation [148]. The expression of both QKI and RBFOX1 for example have direct effects on the splicing of genes enriched for EMT-associated processes such as cell motility, the cytoskeleton and stem cell fate determination and guide specific splicing events of consequence to EMT progression [149]. Examples include the production of a shorter isoform of CD44 that is required to activate AKT during EMT [150] and an exon skipping event that results in the redistribution of FLNB (Filamin B) into the cytoplasm and the subsequent release of the EMT-promoting TF FOXC1 [149].
As with other aspects of EMT, complicated interconnected relationships exist between splicing regulators. For example, the epithelial splicing factor RBM47 (RNA binding motif protein 47) both promotes and antagonizes specific alternate splicing events driven by ESRP [146]. On a similar note, SRSF1-regulated mesenchymal splicing of the Ron tyrosine kinase receptor and the Rac1 GTPase is antagonized by opposing events mediated by the SRSF3 and hnRNPA1 splicing factors in epithelial cells [151, 152]. To add even further complexity, gene regulation during EMT can also operate at the RNA level independently of miRNAs or splicing. PTBP3 (polypyriminine tract binding protein-3) for example promotes EMT through the direct binding and stabilization of the ZEB1 mRNA [153].
Translational regulation
In addition to the regulation of transcription and the influence of microRNAs and alternative splicing on those transcripts, general translational regulatory mechanisms also have a substantial impact upon EMT. YB1 (Y-box binding protein 1) generally suppresses cap-dependent translation on mRNAs but facilitates the cap-independent translation of a subset of mRNAs, including EMT-promoting TFs (SNAI1 ZEB2, TWIST1, LEF1 and TCF4) [154] which are preferentially translated from internal ribosome entry sites (IRES) formed from stem-loop structures within their 5’UTRs [154, 155]. Depletion of the translation initiation factor eIF3e similarly promotes the preferential expression of key EMT TFs [156].
Cap-independent translation can be further aided by N6-methyladeosine (m6A) base modification, with m6A modified mRNAs able to be translated in the absence of eIF4E [157, 158]. m6A modification is the most abundant RNA modification, with functional effects regulated by dynamic interactions among associated methyltransferases (“writers”), demethylases (“erasers”), and binding proteins (“readers”) [159]. TGF-β-induced EMT is inhibited in cells that have reduced expression of the METTL3 “writer”, with lower m6A modification of the TGF-β1 mRNA resulting in lower TGF-β protein production and reduced secretion [160]. Reduced METTL3 expression also downregulated SNAI1 among a group of mRNAs that become m6A-modified during EMT and which is enriched for transcripts encoding proteins related to migration and adherens junctions [161]. In the case of SNAI1, widespread m6A modification throughout the translated region enhanced translational elongation via interaction with YTHDF1, a “reader” that recognizes m6A modified mRNA and recruits the eEF2 translation elongation factor.
Post-translational modification
EMT TFs are also subject to regulatory mechanisms at the post-translational level of which the best characterized is SNAI1, an unstable protein that is rapidly induced in many EMT systems [162]. SNAI1 phosphorylation by CK1 (casein kinase 1), CK2 or DYRK2 (dual-specificity tyrosine-phosphorylation-regulated kinase) primes phosphorylation by GSK3β (glycogen synthase kinase 3β) to create a recognition site for βTCRP, an E3-ubiquitin ligase that leads to SNAIL degradation [163, 164]. Alternately, the phosphorylation of SNAI1 by PAK1 (p21 activated kinase 1) and ATM (ataxia telangiectasia mutated) kinases increase protein stability [14, 165, 166]. Other post-translationally regulated mechanisms of proteasomal degradation have been reported for SNAIL and other core EMT TFs [167,168,169,170,171,172,173,174,175,176]. Phosphorylation can also guide sub-cellular localization. PKD1 (protein kinase D1)-mediated phosphorylation of SNAI1 for example promotes its nuclear export and mutation of the SNAI1 phosphorylation site promotes mesenchymal-like features [168]. In contrast, the nuclear phosphorylation of a different site in SNAI1 by LATS2 (large tumour suppressor kinase-2) promotes EMT by increasing its nuclear retention and stabilization [177]. PC2 (Polycomb protein 2)-mediated sumoylation of ZEB2 on the other hand does not affect protein localization, but instead affects transcriptional activity with sumoylation disrupting the interaction of ZEB2 with the CtBP co-repressor, thus relieving repression of the CDH1 promoter [178].
A case study in complexity: ZEB1
There are two aspects to the notion of extensive, interconnected gene regulatory networks. One is illustrated by the discussion above and the sheer volume of different regulators that have been implicated in the process. Indeed, if the criteria for EMT/MET are taken as a change in marker gene expression along with a phenotypic change to cell morphology or motility after perturbation of the levels of an EMT regulator, at least 300 different TFs and miRNAs have been reported to regulate this process. The other aspect of complex regulation is the multiple levels at which any one of these regulators are connected to others within the network. Thus, examination of the true number, roles and significance of EMT regulators is informed by both the number of participant genes and by their interconnectedness (which operates across different levels of gene expression). As a means of illustrating the complexity of regulation and feedback mechanisms that operate within EMT, we will focus specifically on ZEB1 and examine its regulation by other TFs and non-coding RNAs. In so doing, however, we stress that the situation with this gene is not necessarily more complicated than is the regulation of many other genes within an EMT system, thus highlighting the complex web of control that has evolved.
Regulation of ZEB1
The ZEB1 and ZEB2 proteins, along with members of the SNAIL and TWIST families, constitute core components of the EMT-regulatory network, directly regulating a transcriptional response through interacting with paired E-box motifs within the regulatory regions of genes encoding components of adherens and tight junctions, desomosomes and intermediate filaments [179, 180]. Via the recruitment of histone deacetylases, DNA methyltransferases and components of the SWI/SNF and CtBP co-repressor complexes [181, 182], the ZEBs typically mediate gene repression. However, and in contrast to other core EMT TFs, the ZEBs can also mediate transcriptional activation, recruiting P/CAF and p300 co-activator complexes to promote the expression of mesenchymal genes including as N-cadherin, vimentin, fibronectin and matrix metalloproteases. Conversion from acting as repressors to activators of transcription is brought about via interaction with other proteins including β-catenin and YAP1, effectors of the Wnt and Hippo signaling pathways, respectively [183, 184].
Over the past decade or so, the number of known EMT-regulatory TFs has grown dramatically, though a central pro-mesenchymal function for the ZEBs continues to be reported, often within the context of the miR-200:ZEB co-regulatory loop. Here, members of the miR-200 family (epithelial enforcers) directly bind and downregulate the stability and translation of the ZEB mRNA, whilst the ZEB proteins directly bind and repress the promoters of both miR-200-encoding genomic loci [114, 124, 125, 185]. Illustrating the complexity of the regulation of EMT, even in the context of a single—albeit very important—gene, is that to date, no fewer than 62 different miRNA families have been reported to directly target ZEB1 as indicated by the miRNA-responsive expression of a reporter gene (typically luciferase) fused to the ZEB1-3ʹUTR (Table 1). Such an experiment, considered a gold-standard in the field of miRNA research, is often accompanied by additional measurements of ZEB1 mRNA and protein expression after miRNA perturbation. Even accounting for the reliance on miRNA over-expression in a number of these studies, it is still clear that a gene such as ZEB1 (as well as ZEB2 and members of the SNAIL and TWIST families) are subject to extensive regulatory control.
As previously highlighted, complexity within a regulatory network such as that controlling EMT has multiple levels, first and most obviously the simple number of regulators that have been ascribed this role (such as the > 60 miRNA families reported to directly regulate ZEB1). Complexity is further demonstrated by the frequent participation of ZEB1 in mutual co-regulatory loops with these miRNAs, and by the close association of ZEB1 and these miRNAs with other TFs and miRNAs that themselves also play central roles, building networks motifs of increasing complexity. The reporting of such higher network architecture may be contained within a single study, though the practical limitations of experimental methodologies limit the scope to which this is possible. The heavily studied nature of the field, however, enables a picture of the complex relationships within the network to emerge when considering multiple reports examining smaller subnetworks, motifs and the individual relationships between genes of interest.
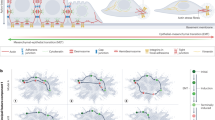
In Fig. 2, such examples are highlighted. In some such examples, the operation of the miR-200:ZEB loop itself can be modulated by other TFs to either bias ZEB (mesenchymal) or miR-200 (epithelial) expression. For example, by directly binding and transactivating the ZEB1 promoter, the Hypoxia Inducible Factors (HIF1a and HIF2a) potentiate EMT (Fig. 2a), which is counteracted by the direct suppression of the HIFs by miR-200 [186, 187]. Alternately, the epithelial enforcer Grainyhead-Like 2 (GRHL2) potentiates MET, promoting the miR-200 arm of the co-regulatory loop by directly promoting transcription of the miR-200 genes, whilst also participating in a direct negative transcriptional loop with ZEB1 (Fig. 2b) [90, 188,189,190,191]. ZEB1 is further modulated by the additional co-regulatory loops in which miR-200 participates. This includes direct reciprocal negative feedback between miR-200 and the SNAIL TF family, whilst the SNAILs, TWIST1 and ETS1 all directly bind and transactivate the ZEB1 promoter. MiR-200 further modulates this regulation, directly targeting ETS1 and both SMAD2/SMAD5 and YWHAB/YWHAG, co-factors for both TWIST and SNAIL, respectively (Fig. 2c) [192, 193]. SNAI1 also participates in a co-regulatory negative feedback loop with miR-34, itself indirectly controlled by ZEB1 via ZEB-mediated transcriptional repression of the miR-34 transactivator, p63 [194]. miR-34 directly targets multiple components of the Wnt signaling pathway, with which both SNAI1 and ZEB1 have direct linkages via the interaction with the Wnt pathway co-activator, β-catenin (CTNNB1) and by direct transcriptional activation by the β-catenin/TCF4 complex (Fig. 2d) [126, 184, 195,196,197,198]. Indeed, this interaction converts ZEB1 from a direct repressor of Wnt pathway targets to an activator as ZEB1 binds TCF4 and swaps co-repressor (Brg1, CtBP) for co-activator (p300) proteins (Fig. 2e) [183]. Perhaps an even more complicated subnetwork applies to the miR-183 ~ 96 ~ 182 cluster, with both ZEB1 and SNAI1/SNAI2 directly repressing transcription of the miRNA host gene whilst multiple miRNAs within the cluster target these same TFs. Interestingly, the miR-183 ~ 96 ~ 182 cluster is also directly transcriptionally activated by B-catenin/Wnt signaling, resulting in multiple complex outcomes downstream as the miRNAs variously target both Wnt pathway activators (LRP6, CTNND1, TCF7L2) and repressors (AXIN2, APC) (Fig. 2f) [199,200,201,202,203,204,205,206].
Regulatory loops incorporating ZEB1 into larger networks involving other TFs and miRNAs. Letter annotations denote supporting PMIDs (A = 26057751; B = 19662677; C = 28899657; D = 22379025; E = 23943797; F = 26887971, 26933170; G = 22370643; H = 25798844; I = 21081489; J = 21317430; K = 21593765, 29259250; L = 22850877; M = 22024162; N = 22421157, 22045851; O = 22080605, 26387539; P = 24277930, 25394902; Q = 23354685; R = 27894095; S = 24289859; T = 31913290; U = 29733821; V = 30070321; W = 31938296)
Examples such as those described above are not intended as an exhaustive catalog of ZEB-associated EMT pathways. Indeed, the number of these interactions would make such a task almost impossible. What it does showcase, however, is that even if one discounts a large volume of studies that are reliant upon over-expression or those in which EMT-associated outcomes are demonstrated by limited means, tremendous complexity is still apparent—even if one just considers a single EMT regulator like ZEB1. This same conclusion would also be drawn from an in-depth focus into the regulation of SNAI1, SNAI2, TWIST1 and any number of dozens of other key TFs for which the sum of evidence supporting their EMT-regulatory capacity is overwhelming.
EMT: why complexity is a necessity
What is known about any system is proportional to the time devoted to its study. Thus, given the importance of EMT to both development and pathology, enormous effort has been dedicated to understanding EMT regulation and function. Even with this in mind, however, it is clear that tremendous complexity has evolved around EMT, illustrated not just by a large number of regulators, but also via the multiple levels of gene regulation at which they operate. By necessity, such complexity is largely ignored in individual studies that seek to uncover roles for specific genes in specific contexts. Here, we consider why such complexity is necessary.
The ubiquity of EMT/MET: multiple contexts
During development, a broad range of stimuli are deployed at different times and in different sites to guide EMT in a diverse range of cells. Therefore, it is perhaps not surprising that a large number of sensors, effectors and modulators have evolved to facilitate EMT across the multiple contexts in which it is activated. The diverse nature of responsive cells is seen by the fact that EMT/MET-like processes, often driven by the same core EMT TFs, still promote the aggressiveness of cancers in tissues of non-directly epithelial or mesenchymal origin such as gliomas (originally derived from primitive neuroectoderm), sarcomas and haematological malignancies (derived from muscle and bone, originally of the embryonic mesoderm) [207].
Another complexity is that EMT itself varies widely across different contexts, both in terms of the range of possible phenotypic outcomes and in the nature of the underlying transcriptomic profiles. For example, the majority of genes that were differentially expressed between cell lines derived from the lung, kidney and breast were unique in response to the same EMT stimulus (TGFβ + TNFα) [208]. Transcriptomic profiling of single-cell lines of ovarian, prostate, breast and lung origin in response to 3 different EMT inducers (TGFβ, EGF, TNFα) also showed little overlap of responsive genes, both between the same stimulus across different cells and between different stimuli in the same cell [209]. It is therefore clear that not only are there divergent EMT pathways between cells, but that multiple EMT pathways are operable in the same cell, triggered by ligands binding different cell surface receptor kinases. Additionally, the promoters of key genes such as CDH1 [210] and ZEB1 [211] can simultaneously display both repressive and active marks, creating a poised bi-valent state which allows rapid on–off cycling and likely contributes to EMT reversibility. Complexity therefore broadly arises from several sources: the requirement for cells with a differing gene expression landscape to still be responsive to EMT, and to facilitate different phenotypic outcomes tailored to the specific context and nature of the stimulus.
Flexibility–reversibility and partial EMT phenotypes
Historically, EMT has been viewed as a binary process whereby cells undergo transformations between epithelial and mesenchymal states, as often defined by the gain and loss of select epithelial (E-cadherin) and mesenchymal (N-cadherin, vimentin) markers. Subsequent mathematical models and biological observations, however, now supported by single-cell RNA sequencing (scRNA-Seq), clearly demonstrate there exists a spectrum of hybrid phenotypes (also referred to as “incomplete”, “intermediate” or “partial” EMT), whereby individual cells co-express both epithelial and mesenchymal markers [212].
Not only have hybrid states been noted across a diverse range of cells, both in cell culture and in vivo (reviewed in [212]), but the existence of a hybrid state itself is of tremendous functional significance as it is tied to the capacity of cells to migrate during both development and cancer and in the promotion of stemness properties. Hybrid E/M phenotypes allow collective cell migration by maintaining adhesion between neighbouring cells whilst decreasing apico-basal polarity, thus increasing the motility of the leading cells. Collective migration is used during embryonic development such as in the branching morphogenesis of the mammary gland or the sprouting angiogenesis of endothelial cells [213, 214]. It is also employed in the adult, both in essential processes such as wound healing [215] and in pathologies for which it has gained particular attention. Fibrotic renal tubular epithelial cells for example display a hybrid EMT phenotype [216,217,218], as do cells at the invasive front of tumours which corresponds to poor survival across many tumour types [24, 219]. Circulating-tumour cells (CTCs) that are associated with a diverse array of cancers also display a hybrid phenotype [35, 220,221,222,223] and the presence of a hybrid state is more closely associated with a poor clinical outcome than are fully epithelial or mesenchymal features [220, 224,225,226,227].
Initially, EMT was proposed to increase stemness [228, 229], however, it was subsequently found that cells that become locked in an exclusively mesenchymal state actually lose their stem-like properties [101, 230] and it is in fact the hybrid state that creates a “stemness window” [231, 232]. Cells displaying such hybrid features exhibit increased tumourigenic capacity [233] and the ectopic expression of EMT TFs enhances the formation of secondary tumours upon transplantation [228, 229]. Growing evidence also links the hybrid E/M phenotype with the resistance to therapy [43, 234,235,236,237], further suggesting that targeting hybrid E/M cells may be a productive focus for therapeutic strategies.
The mathematical modelling that was initially employed to predict stable intermediate states (reviewed in [238, 239]) has been superseded by single-cell sequencing which suggests that waves of continuous gene expression give rise to a myriad of intermediate phenotypes [219, 240,241,242]. The capacity of cells to reside within such hybrid states is supported by phenotypic stability factors (PSFs); genes expressed in hybrid E/M phenotypes that counteract the full transition by regulating core EMT regulators [232, 238, 239, 243]. The first such PSFs to be predicted and experimentally validated are OVOL2 and GRHL2, TFs known to induce MET or halt EMT in a context-dependent manner by suppressing several EMT promoting TFs such as ZEB1 with which they form mutually inhibitory loops of regulation [89, 90, 190, 244,245,246,247]. Subsequently, the list of factors to be given the PSF designation has grown. For example, miRNAs such as miR-145 and miR-129 have been described as PSFs through the opposing roles they play against ZEB2 [248] and TWIST1 [249] respectively, whilst NRF2 contributes to a hybrid E/M state by suppressing SNAIL [250].
It may be that certain factors specifically function as PSFs. NFATc for example induces both epithelial (E-cadherin, miR-200) and mesenchymal (ZEB1) genes and thereby stabilises a hybrid E/M phenotype [251]. Given the interdependent nature of gene regulation, however, it could be argued essentially all system components contribute a PSF role. Even E-cadherin (CDH1) for example could be regarded in this manner, sequestering B-catenin which in turn prevents the transcriptional activation of ZEB and thereby, prevents ZEB’s inhibitory role on the CDH1 promoter [252]. We would argue therefore that for the most part, PSFs should be thought of less as specific hybrid maintenance factors, and more simply as components of the large and overarching genetic networks through which the E/M status of a cell is derived as a result of the multitude of opposing signaling outcomes (Fig. 3). This notion of multiple PSFs functioning as small parts within wider networks may also be a more helpful way to approach complex networks, given the tendency in papers to ascribe major consequences to single effectors which both comes from the publication-incentivised over-interpretation of results and the over-simplification of gene regulation that necessarily follows.
Along with the growing number of studies that demonstrate “non-canonical” TFs (such as HEY1, FOXO3 and FOXA1) can also induce EMT independently of the canonical EMT TF core, the number of potential PSFs that interact with them will also grow, further enlarging the complexity of EMT regulation [239]. It is yet unknown to what extent non-core TFs can drive EMT and it may be that their contribution is larger than generally recognised. ScRNA-Seq for example identified the widespread hybrid characteristics of developing intestinal, lung and liver cells despite their very low expression of SNAIL, ZEB and TWIST family genes [240]. By employing additional PSFs to also regulate these regulators, the EMT system further increases its information capacity, meaning the system can reside in multiple distinct states for specific purposes, further fine-tuning traits associated with motility and stemness.
Noise reduction, buffering and inbuilt safety
The requirement that many types of cells be responsive to EMT is a likely driver of the large number of stimuli that are capable of promoting EMT. Cells, however, are required to balance responsiveness with blocking inadvertent activation, as this could have serious consequences, including promoting the metastasis of cancer. The potential for spurious induction is heightened by the observation that even a common metabolite such as oxalate can promote EMT [73], raising the question of how many other metabolic by-products to which cells are frequently exposed could also act as triggers? In addition to exposure to a broad range of external stimuli, random fluctuations in the expression level of individual genes within the EMT network could also propagate through the system if such gene expression noise impacts upon key targets such as regulatory TFs. If these fluctuations occur around a critical threshold of TF abundance, for example, the noise from initially minor variations could be propagated to result in radical transcriptomic (and phenotypic) changes. The need to buffer such transcriptomic noise necessitates the evolution of more complex regulatory programs and higher scales of network architecture. The size and complexity of the gene regulatory networks (GRNs) associated with EMT reflect the importance of correct regulation of the process, as well as those additional features such as the diversity of contexts in which EMT must operate, the need for reversibility and the specific importance of hybrid states.
The architectures of GRNs are built from circuits and sub-circuits of interacting genes that mediate specific responses [253]. One such example of relevance is the distinction of 5 EMT sub-circuits (controlling basement remodeling, motility, apical constriction, apical-basal polarity and de-adhesion) operational during sea urchin development [254]. Even in this relatively simple system (primary mesenchyme cells; the first cells in sea urchin development to undergo EMT) complex networks exist in which there is the absence of any single master regulatory TF. This is because at least 13 different TFs are required for the completion of EMT, though no single TF is required for each of the 5 sub-circuits [254]. Single-cell sequencing that follows the progression of EMT has also noted waves of gene expression and series of discrete transcriptional events, suggesting EMT is a multistep process even though it presents as a continuous gradient of gene expression without discernable boundaries between hybrid states [209].
As briefly discussed earlier, complex networks include smaller recurring circuits called “network motifs” which can be broadly divided into two categories: feedback and feedforward [255]. Positive feedback loops often underlie developmental switches; for example, a TF promoting its own expression to facilitate an “all or none” outcome. If that same TF, however, were to induce a repressor of its own expression, a negative feedback loop such as this would limit strong changes. Other types of negative feedback, however, are conducive to molecular switches. The reciprocal feedback loop between ZEB and miR-200 is one such example, where a TF directly represses a miRNA that itself targets the TF. Feedforward motifs on the other hand are based on regulators that act both directly and indirectly on their downstream targets. Multiple outcomes are therefore possible depending upon the nature of the motif. Irrespective of specific network motifs, one overarching principal is that GRNs that incorporate positive and negative feedback increase their potential to control the effects of noise, buffering its impact on gene expression [255,256,257,258].
The buffering of biological noise, and the capacity to directly modulate the activity of TFs, make miRNAs ideally suited to the regulation of complex processes. This would explain the association of many miRNAs with EMT, well beyond the best-established examples such as miR-200, miR-203 and miR-34. MiRNA-TF motifs are represented in biological networks at a much higher rate than would be expected by chance; both in a directly reciprocal manner and where a TF positively regulates both a miRNA and a target gene that the same miRNA also negatively regulates [121]. This would explain the seemingly contradictory observation that the expression of miRNAs and their targets are often positively correlated [259, 260]. Experimentally, the capacity of a miRNA to buffer noise was demonstrated using an artificial reporter system consisting of an inducible, self-regulatory TF whose expression level controls an on/off “toggle-switch” phenotype, coupled with a miRNA that targets the TF. When present, the miRNA conferred robustness and enabled the cell to maintain its state though when absent, a dramatic increase in protein noise level caused the cell to randomly switch between states [261]. On a transcriptome-level scale, noise from lowly expressed genes is buffered by miRNAs and genes regulated by multiple miRNAs show greater noise reduction [262]. MicroRNAs also participate in the widespread buffering of transcriptomic noise in EMT systems [263] and the 3ʹUTRs from genes with variably active promoters are more frequently targeted by miRNAs than are the 3ʹUTRs of genes of low transcriptional noise [264].
The requirement for multiple direct regulators may also be a function of the contribution of any one regulator, even a core EMT TF, being insufficient in itself to sway the phenotype in isolation. Thus, a comparatively “weak” effect that is mediated by any individual regulator provides an inbuilt safety mechanism, minimizing the genetic noise that could result from dysregulation of any single factor in isolation. Such a model is consistent with the successive waves of TF expression that is reported after exposure to EMT-inducing stimuli, with the expression of later regulators being dependent upon the upregulation of more rapid responders and their co-operative actions.
Concluding remarks
EMT encompasses a broad range of processes that are measurable by a number of genetic markers and phenotypic outcomes. It is therefore inherently difficult to establish a minimal evidential threshold to define an EMT regulator (despite recent attempts [4]) which allows one to question the importance of many proposed regulators that are yet to be substantiated by multiple laboratories, or for which the evidence is dependent upon exogenous expression or the measurement of limited markers or phenotypes. Even with this in mind, however, it is clear that EMT/MET is subject to regulation by many dozens (and likely hundreds) of different TFs and miRNAs, themselves subject to additional levels of control via splicing and post-translational modification.
Complexity is apparent with the regulation of any genetic process, though may take on special importance with EMT due to several factors. These include the requirement that multiple types of cells during development and in the adult be capable of E/M plasticity, and the hybrid nature of the process itself as cells balance and combine various features of epithelial and mesenchymal states to facilitate specific outcomes. For the researcher, multiple factors must therefore be considered if one is to take a global view of a complex regulatory process such as EMT. For example, how many phenotypes are associated with EMT? What are the tipping points that restrict reversibility and if a reversal of phenotype occurs, are the molecular pathways involved simply reversed? Further, how is the buffering of gene expression balanced with the capacity for phenotypic change when both of these processes are facilitated by a common pool of potential regulators? Mathematical tools to assist the modelling of such complexity are available [265].
By bolting on feedback and feedforward loops (within which TFs drive genetic programs with miRNAs providing a critical regulatory layer), the information capacity of the system (how many states a system can exist in) is increased, facilitating hybrid states whilst providing additional noise reduction and buffering capacity. In this light, many (if not all) regulators can be viewed as phenotypic stability factors (PSFs)—important contributors that at their endogenous levels do not necessarily lock a phenotype at either end of the E/M spectrum, but rather, that balance opposing effects of other regulators as phenotypes are stabilised along an E/M continuum.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Abbreviations
- EMT:
-
Epithelial-mesenchymal transition
- MET:
-
Mesenchymal-epithelial transition
- EMP:
-
Epithelial-mesenchymal plasticity
- miRNA:
-
MicroRNA
- TF:
-
Transcription factor
- GRN:
-
Gene regulatory network
- PSF:
-
Phenotypic stability factor
- scRNA-Seq:
-
Single-cell RNA sequencing
- CTC:
-
Circulating tumour cell
References
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196
Nieto MA, Huang RY, Jackson RA, Thiery JP (2016) Emt: 2016. Cell 166:21–45
Zhang Y, Weinberg RA (2018) Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med 12:361–373
Yang J et al (2020) Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 21:341–352
Greenburg G, Hay ED (1982) Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J Cell Biol 95:333–339
Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J (2005) The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell 8:179–192
Dale JK et al (2006) Oscillations of the snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev Cell 10:355–366
Timmerman LA et al (2004) Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 18:99–115
Haensel D, Dai X (2018) Epithelial-to-mesenchymal transition in cutaneous wound healing: where we are and where we are heading. Dev Dyn 247:473–480
Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG (2002) Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest 110:341–350
Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA (2006) Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci USA 103:13180–13185
Flier SN, Tanjore H, Kokkotou EG, Sugimoto H, Zeisberg M, Kalluri R (2010) Identification of epithelial to mesenchymal transition as a novel source of fibroblasts in intestinal fibrosis. J Biol Chem 285:20202–20212
Aiello NM, Kang Y (2019) Context-dependent EMT programs in cancer metastasis. J Exp Med 216:1016–1026
De Craene B, Berx G (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13:97–110
Tsubakihara Y, Moustakas A (2018) Epithelial-mesenchymal transition and metastasis under the control of transforming growth factor beta. Int J Mol Sci 19:3672
Williams ED, Gao D, Redfern A, Thompson EW (2019) Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat Rev Cancer 19:716–732
Bracken CP, Scott HS, Goodall GJ (2016) A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 17:719–732
Lu W, Kang Y (2019) Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell 49:361–374
Celia-Terrassa T, Kang Y (2016) Distinctive properties of metastasis-initiating cells. Genes Dev 30:892–908
Futterman MA, Garcia AJ, Zamir EA (2011) Evidence for partial epithelial-to-mesenchymal transition (pEMT) and recruitment of motile blastoderm edge cells during avian epiboly. Dev Dyn 240:1502–1511
Grigore AD, Jolly MK, Jia D, Farach-Carson MC, Levine H (2016) Tumor budding: the name is EMT. Partial EMT. J Clin Med 5:51
Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, Onuchic JN, Levine H (2015) Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol 5:155
Saitoh M (2018) Involvement of partial EMT in cancer progression. J Biochem 164:257–264
Shamir ER et al (2014) Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J Cell Biol 204:839–856
Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR (2008) Cadherin switching. J Cell Sci 121:727–735
Brabletz T, Jung A, Hermann K, Gunther K, Hohenberger W, Kirchner T (1998) Nuclear overexpression of the oncoprotein beta-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol Res Pract 194:701–704
Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, Knuechel R, Kirchner T (2001) Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA 98:10356–10361
Bronsert P et al (2014) Cancer cell invasion and EMT marker expression: a three-dimensional study of the human cancer-host interface. J Pathol 234:410–422
Kunita A et al (2018) Inflammatory cytokines induce podoplanin expression at the tumor invasive front. Am J Pathol 188:1276–1288
Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ (2013) Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia 15:180–191
Zhao Z et al (2016) In vivo visualization and characterization of epithelial-mesenchymal transition in breast tumors. Cancer Res 76:2094–2104
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S (2009) Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 11:R46
Hyun KA, Koo GB, Han H, Sohn J, Choi W, Kim SI, Jung HI, Kim YS (2016) Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 7:24677–24687
Lapin M, Tjensvoll K, Oltedal S, Javle M, Smaaland R, Gilje B, Nordgard O (2017) Single-cell mRNA profiling reveals transcriptional heterogeneity among pancreatic circulating tumour cells. BMC Cancer 17:390
Yu M et al (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339:580–584
Husemann Y et al (2008) Systemic spread is an early step in breast cancer. Cancer Cell 13:58–68
Raimondi C et al (2011) Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat 130:449–455
Jang MH, Kim HJ, Kim EJ, Chung YR, Park SY (2015) Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum Pathol 46:1267–1274
Ryu HS et al (2012) Overexpression of epithelial-mesenchymal transition-related markers according to cell dedifferentiation: clinical implications as an independent predictor of poor prognosis in cholangiocarcinoma. Hum Pathol 43:2360–2370
Olmeda D, Montes A, Moreno-Bueno G, Flores JM, Portillo F, Cano A (2008) Snai1 and Snai2 collaborate on tumor growth and metastasis properties of mouse skin carcinoma cell lines. Oncogene 27:4690–4701
Spaderna S et al (2008) The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 68:537–544
Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ (2008) Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol 10:295–305
Yin T, Wang C, Liu T, Zhao G, Zha Y, Yang M (2007) Expression of snail in pancreatic cancer promotes metastasis and chemoresistance. J Surg Res 141:196–203
Jensen C, Teng Y (2020) Is it time to start transitioning from 2d to 3d cell culture? Front Mol Biosci 7:33
Groger CJ, Grubinger M, Waldhor T, Vierlinger K, Mikulits W (2012) Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS ONE 7:e51136
Jung AR, Jung CH, Noh JK, Lee YC, Eun YG (2020) Epithelial-mesenchymal transition gene signature is associated with prognosis and tumor microenvironment in head and neck squamous cell carcinoma. Sci Rep 10:3652
Parsana P, Amend SR, Hernandez J, Pienta KJ, Battle A (2017) Identifying global expression patterns and key regulators in epithelial to mesenchymal transition through multi-study integration. BMC Cancer 17:447
Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, Thiery JP (2014) Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med 6:1279–1293
Taube JH et al (2010) Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA 107:15449–15454
Vasaikar SV et al (2021) EMTome: a resource for pan-cancer analysis of epithelial-mesenchymal transition genes and signatures. Br J Cancer 124:259–269
Weiss A, Attisano L (2013) The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol 2:47–63
Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA (2009) Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest 119:1438–1449
Kruithof BP, Duim SN, Moerkamp AT, Goumans MJ (2012) TGFbeta and BMP signaling in cardiac cushion formation: lessons from mice and chicken. Differentiation 84:89–102
Mercado-Pimentel ME, Runyan RB (2007) Multiple transforming growth factor-beta isoforms and receptors function during epithelial-mesenchymal cell transformation in the embryonic heart. Cells Tissues Organs 185:146–156
Sauka-Spengler T, Bronner-Fraser M (2008) A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol 9:557–568
Nawshad A, LaGamba D, Hay ED (2004) Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT). Arch Oral Biol 49:675–689
Willis BC, Borok Z (2007) TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol 293:L525–L534
Gressner AM, Weiskirchen R, Breitkopf K, Dooley S (2002) Roles of TGF-beta in hepatic fibrosis. Front Biosci 7:d793-807
Schnaper HW, Hayashida T, Hubchak SC, Poncelet AC (2003) TGF-beta signal transduction and mesangial cell fibrogenesis. Am J Physiol Renal Physiol 284:F243–F252
Grande M, Franzen A, Karlsson JO, Ericson LE, Heldin NE, Nilsson M (2002) Transforming growth factor-beta and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes. J Cell Sci 115:4227–4236
Canadas I et al (2014) High circulating hepatocyte growth factor levels associate with epithelial to mesenchymal transition and poor outcome in small cell lung cancer patients. Oncotarget 5:5246–5256
Strutz F, Zeisberg M, Ziyadeh FN, Yang CQ, Kalluri R, Muller GA, Neilson EG (2002) Role of basic fibroblast growth factor-2 in epithelial-mesenchymal transformation. Kidney Int 61:1714–1728
Yang AD et al (2006) Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res 66:46–51
Graham TR et al (2008) Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res 68:2479–2488
Fernando RI, Castillo MD, Litzinger M, Hamilton DH, Palena C (2011) IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res 71:5296–5306
Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP (2004) Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J 23:1155–1165
Katoh Y, Katoh M (2008) Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int J Mol Med 22:271–275
Kim K, Lu Z, Hay ED (2002) Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol Int 26:463–476
Liebner S, Cattelino A, Gallini R, Rudini N, Iurlaro M, Piccolo S, Dejana E (2004) Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J Cell Biol 166:359–367
Sun S et al (2009) Hypoxia-inducible factor-1alpha induces twist expression in tubular epithelial cells subjected to hypoxia, leading to epithelial-to-mesenchymal transition. Kidney Int 75:1278–1287
Tam SY, Wu VWC, Law HKW (2020) Hypoxia-induced epithelial-mesenchymal transition in cancers: HIF-1alpha and beyond. Front Oncol 10:486
Heise RL, Stober V, Cheluvaraju C, Hollingsworth JW, Garantziotis S (2011) Mechanical stretch induces epithelial-mesenchymal transition in alveolar epithelia via hyaluronan activation of innate immunity. J Biol Chem 286:17435–17444
Convento MB, Pessoa EA, Cruz E, da Gloria MA, Schor N, Borges FT (2017) Calcium oxalate crystals and oxalate induce an epithelial-to-mesenchymal transition in the proximal tubular epithelial cells: contribution to oxalate kidney injury. Sci Rep 7:45740
Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2:84–89
Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2:76–83
Hajra KM, Chen DY, Fearon ER (2002) The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res 62:1613–1618
Comijn J et al (2001) The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell 7:1267–1278
Vandewalle C et al (2005) SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res 33:6566–6578
Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C (2010) The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest 120:533–544
Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A (2001) A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem 276:27424–27431
Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J (2007) Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res 67:7184–7193
Yang J et al (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927–939
Ocana OH et al (2012) Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22:709–724
Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi DC, Weinberg RA (2006) The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc Natl Acad Sci USA 103:18969–18974
Sobrado VR, Moreno-Bueno G, Cubillo E, Holt LJ, Nieto MA, Portillo F, Cano A (2009) The class I bHLH factors E2–2A and E2–2B regulate EMT. J Cell Sci 122:1014–1024
McCoy EL, Iwanaga R, Jedlicka P, Abbey NS, Chodosh LA, Heichman KA, Welm AL, Ford HL (2009) Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J Clin Invest 119:2663–2677
Mani SA et al (2007) Mesenchyme forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA 104:10069–10074
Tiwari N et al (2013) Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23:768–783
Roca H et al (2013) Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS ONE 8:e76773
Cieply B, Riley P, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM (2012) Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res 72:2440–2453
Chang CJ et al (2011) p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 13:317–323
Chakrabarti R et al (2012) Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol 14:1212–1222
Chou CC, Lee KH, Lai IL, Wang D, Mo X, Kulp SK, Shapiro CL, Chen CS (2014) AMPK reverses the mesenchymal phenotype of cancer cells by targeting the Akt-MDM2-Foxo3a signaling axis. Cancer Res 74:4783–4795
Song Y, Washington MK, Crawford HC (2010) Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res 70:2115–2125
Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A (2008) HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem 283:33437–33446
Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A (2006) Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol 174:175–183
Gregory PA et al (2011) An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell 22:1686–1698
Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, de Herreros AG (2011) Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem 286:12024–12032
Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J (2011) Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res 71:245–254
Krebs AM et al (2017) The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 19:518–529
Tran HD, Luitel K, Kim M, Zhang K, Longmore GD, Tran DD (2014) Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res 74:6330–6340
Xu Y, Lee DK, Feng Z, Xu Y, Bu W, Li Y, Liao L, Xu J (2017) Breast tumor cell-specific knockout of Twist1 inhibits cancer cell plasticity, dissemination, and lung metastasis in mice. Proc Natl Acad Sci USA 114:11494–11499
Park SY et al (2015) Combinatorial TGF-beta attenuation with paclitaxel inhibits the epithelial-to-mesenchymal transition and breast cancer stem-like cells. Oncotarget 6:37526–37543
Wang J et al (2017) Snail determines the therapeutic response to mTOR kinase inhibitors by transcriptional repression of 4E-BP1. Nat Commun 8:2207
Chiu LY, Hsin IL, Yang TY, Sung WW, Chi JY, Chang JT, Ko JL, Sheu GT (2017) The ERK-ZEB1 pathway mediates epithelial-mesenchymal transition in pemetrexed resistant lung cancer cells with suppression by vinca alkaloids. Oncogene 36:242–253
Sayan AE et al (2009) SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proc Natl Acad Sci USA 106:14884–14889
Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J (2009) A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15:489–500
Stemmler MP, Eccles RL, Brabletz S, Brabletz T (2019) Non-redundant functions of EMT transcription factors. Nat Cell Biol 21:102–112
Caramel J et al (2013) A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24:466–480
Postigo AA (2003) Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J 22:2443–2452
Postigo AA, Depp JL, Taylor JJ, Kroll KL (2003) Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J 22:2453–2462
Skrypek N, Goossens S, De Smedt E, Vandamme N, Berx G (2017) Epithelial-to-mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet 33:943–959
Feldker N et al (2020) Genome-wide cooperation of EMT transcription factor ZEB1 with YAP and AP-1 in breast cancer. EMBO J 39:e103209
Gregory PA et al (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10:593–601
Wyatt L, Wadham C, Crocker LA, Lardelli M, Khew-Goodall Y (2007) The protein tyrosine phosphatase Pez regulates TGFbeta, epithelial-mesenchymal transition, and organ development. J Cell Biol 178:1223–1235
Cui Q, Yu Z, Purisima EO, Wang E (2006) Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol 2:46
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–798
Gerstein MB et al (2012) Architecture of the human regulatory network derived from ENCODE data. Nature 489:91–100
Shalgi R, Lieber D, Oren M, Pilpel Y (2007) Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 3:e131
Su WL, Kleinhanz RR, Schadt EE (2011) Characterizing the role of miRNAs within gene regulatory networks using integrative genomics techniques. Mol Syst Biol 7:490
Tsang J, Zhu J, van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26:753–767
Alon U (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8:450–461
Becskei A, Serrano L (2000) Engineering stability in gene networks by autoregulation. Nature 405:590–593
Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68:7846–7854
Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 9:582–589
Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H (2011) miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 10:4256–4271
Moes M, Le Bechec A, Crespo I, Laurini C, Halavatyi A, Vetter G, Del Sol A, Friederich E (2012) A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PLoS ONE 7:e35440
Liu YN et al (2013) MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 32:296–306
Fazilaty H, Rago L, Kass Youssef K, Ocana OH, Garcia-Asencio F, Arcas A, Galceran J, Nieto MA (2019) A gene regulatory network to control EMT programs in development and disease. Nat Commun 10:5115
Meyer-Schaller N et al (2019) A hierarchical regulatory landscape during the multiple stages of EMT. Dev Cell 48:539–553
Diepenbruck M, Tiede S, Saxena M, Ivanek R, Kalathur RKR, Luond F, Meyer-Schaller N, Christofori G (2017) miR-1199-5p and Zeb1 function in a double-negative feedback loop potentially coordinating EMT and tumour metastasis. Nat Commun 8:1168
Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27:91–105
Bracken CP et al (2014) Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. EMBO J 33:2040–2056
Hoefert JE, Bjerke GA, Wang D, Yi R (2018) The microRNA-200 family coordinately regulates cell adhesion and proliferation in hair morphogenesis. J Cell Biol 217:2185–2204
Jurmeister S et al (2012) MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol 32:633–651
Pillman KA et al (2019) Extensive transcriptional responses are co-ordinated by microRNAs as revealed by exon-intron split analysis (EISA). Nucleic Acids Res 47:8606–8619
Gosline SJ et al (2016) Elucidating MicroRNA regulatory networks using transcriptional, post-transcriptional, and histone modification measurements. Cell Rep 14:310–319
Neumann DP, Goodall GJ, Gregory PA (2018) Regulation of splicing and circularisation of RNA in epithelial mesenchymal plasticity. Semin Cell Dev Biol 75:50–60
Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB (2011) An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7:e1002218
Warzecha CC et al (2010) An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J 29:3286–3300
Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP (2009) ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33:591–601
Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ (2008) A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem 283:18344–18354
Horiguchi K et al (2012) TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene 31:3190–3201
Reinke LM, Xu Y, Cheng C (2012) Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem 287:36435–36442
Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J (2014) The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene 33:1082–1092
Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, Xing Y, Carstens RP (2016) Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol Cell Biol 36:1704–1719
Venables JP et al (2013) RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol 33:396–405
Conn SJ et al (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134
Li J et al (2018) An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. Elife 7:e37184
Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C (2011) CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest 121:1064–1074
Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G (2013) HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res 41:8665–8679
Goncalves V, Matos P, Jordan P (2009) Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet 18:3696–3707
Hou P, Li L, Chen F, Chen Y, Liu H, Li J, Bai J, Zheng J (2018) PTBP3-mediated regulation of zeb1 mrna stability promotes epithelial-mesenchymal transition in breast cancer. Cancer Res 78:387–398
Evdokimova V et al (2009) Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15:402–415
Desnoyers G, Frost LD, Courteau L, Wall ML, Lewis SM (2015) Decreased eIF3e Expression can mediate epithelial-to-mesenchymal transition through activation of the TGFbeta signaling pathway. Mol Cancer Res 13:1421–1430
Gillis LD, Lewis SM (2013) Decreased eIF3e/Int6 expression causes epithelial-to-mesenchymal transition in breast epithelial cells. Oncogene 32:3598–3605
Bera A, Lewis SM (2020) Regulation of epithelial-to-mesenchymal transition by alternative translation initiation mechanisms and its implications for cancer metastasis. Int J Mol Sci 21:4075
Meyer KD et al (2015) 5’ UTR m(6)A promotes cap-independent translation. Cell 163:999–1010
Meyer KD, Jaffrey SR (2014) The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 15:313–326
Li J, Chen F, Peng Y, Lv Z, Lin X, Chen Z, Wang H (2020) N6-methyladenosine regulates the expression and secretion of TGFbeta1 to affect the epithelial-mesenchymal transition of cancer cells. Cells 9:296
Lin X et al (2019) RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of snail. Nat Commun 10:2065
Baulida J, Diaz VM, Herreros AG (2019) Snail1: a transcriptional factor controlled at multiple levels. J Clin Med 8:757
Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ (2005) Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem 280:11740–11748
Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC (2004) Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 6:931–940
Sun M et al (2012) Activation of the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell Biol 4:304–315
Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R (2005) Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail’s subcellular localization and functions. Cancer Res 65:3179–3184
Chen A, Wong CS, Liu MC, House CM, Sceneay J, Bowtell DD, Thompson EW, Moller A (2015) The ubiquitin ligase Siah is a novel regulator of Zeb1 in breast cancer. Oncotarget 6:862–873
Du C, Zhang C, Hassan S, Biswas MH, Balaji KC (2010) Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res 70:7810–7819
Hong J, Zhou J, Fu J, He T, Qin J, Wang L, Liao L, Xu J (2011) Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res 71:3980–3990
Li CW et al (2016) AKT1 inhibits epithelial-to-mesenchymal transition in breast cancer through phosphorylation-dependent twist1 degradation. Cancer Res 76:1451–1462
Lim SO, Kim H, Jung G (2010) p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS Lett 584:2231–2236
Lin Y et al (2017) Stabilization of the transcription factors slug and twist by the deubiquitinase dub3 is a key requirement for tumor metastasis. Oncotarget 8:75127–75140
Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, Diaz VM (2014) Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res 42:1079–1094
Wang SP et al (2009) p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 11:694–704
Wang WL et al (2015) Slug is temporally regulated by cyclin E in cell cycle and controls genome stability. Oncogene 34:1116–1125
Zhou Z et al (2017) USP51 promotes deubiquitination and stabilization of ZEB1. Am J Cancer Res 7:2020–2031
Zhang K, Rodriguez-Aznar E, Yabuta N, Owen RJ, Mingot JM, Nojima H, Nieto MA, Longmore GD (2012) Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J 31:29–43
Long J, Zuo D, Park M (2005) Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J Biol Chem 280:35477–35489
Caramel J, Ligier M, Puisieux A (2018) Pleiotropic roles for ZEB1 in cancer. Cancer Res 78:30–35
Vandewalle C, Van Roy F, Berx G (2009) The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci 66:773–787
Postigo AA, Dean DC (1999) ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA 96:6683–6688
Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A (2010) ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 29:3490–3500
Lehmann W et al (2016) ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun 7:10498
Sanchez-Tillo E, de Barrios O, Valls E, Darling DS, Castells A, Postigo A (2015) ZEB1 and TCF4 reciprocally modulate their transcriptional activities to regulate Wnt target gene expression. Oncogene 34:5760–5770
Park SM, Gaur AB, Lengyel E, Peter ME (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22:894–907
Shang Y, Chen H, Ye J, Wei X, Liu S, Wang R (2017) HIF-1alpha/Ascl2/miR-200b regulatory feedback circuit modulated the epithelial-mesenchymal transition (EMT) in colorectal cancer cells. Exp Cell Res 360:243–256
Zhang W et al (2015) HIF-1alpha promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS ONE 10:e0129603
Chen W et al (2016) Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis 37:500–510
Chung VY et al (2016) GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep 6:19943
Cieply B, Farris J, Denvir J, Ford HL, Frisch SM (2013) Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res 73:6299–6309
Somarelli JA et al (2016) Mesenchymal-epithelial transition in sarcomas is controlled by the combinatorial expression of microRNA 200s and GRHL2. Mol Cell Biol 36:2503–2513
Chan YC, Khanna S, Roy S, Sen CK (2011) miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem 286:2047–2056
Perdigao-Henriques R, Petrocca F, Altschuler G, Thomas MP, Le MT, Tan SM, Hide W, Lieberman J (2016) miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 35:158–172
Ahn YH et al (2012) ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest 122:3170–3183
Kim NH et al (2011) p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Sig 4:ra71
Kim NH et al (2011) A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol 195:417–433
Sanchez-Tillo E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A (2011) beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci USA 108:19204–19209
Xiao YY et al (2019) ZEB1 promotes invasion and metastasis of endometrial cancer by interacting with HDGF and inducing its transcription. Am J Cancer Res 9:2314–2330
Chen C, Xiang H, Peng YL, Peng J, Jiang SW (2014) Mature miR-183, negatively regulated by transcription factor GATA3, promotes 3T3-L1 adipogenesis through inhibition of the canonical Wnt/beta-catenin signaling pathway by targeting LRP6. Cell Sig 26:1155–1165
Chen D, Li SG, Chen JY, Xiao M (2018) MiR-183 maintains canonical Wnt signaling activity and regulates growth and apoptosis in bladder cancer via targeting AXIN2. Eur Rev Med Pharmacol Sci 22:4828–4836
Gao XH, Zhang YL, Zhang ZY, Guo SS, Chen XB, Guo YZ (2020) MicroRNA-96-5p represses breast cancer proliferation and invasion through Wnt/beta-catenin signaling via targeting CTNND1. Sci Rep 10:44
Leung WK, He M, Chan AW, Law PT, Wong N (2015) Wnt/beta-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett 362:97–105
Li H, Ma Y, Chen B, Shi J (2018) miR-182 enhances acute kidney injury by promoting apoptosis involving the targeting and regulation of TCF7L2/Wnt/beta-catenins pathway. Eur J Pharmacol 831:20–27
Liu X, Li H, Wu G, Cui S (2018) miR-182 promotes cell proliferation and invasion by inhibiting APC in melanoma. Int J Clin Exp Pathol 11:1900–1908
Tang X et al (2014) Glycogen synthase kinase 3 beta inhibits microRNA-183-96-182 cluster via the beta-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic Acids Res 42:2988–2998
Wang J, Wang X, Li Z, Liu H, Teng Y (2014) MicroRNA-183 suppresses retinoblastoma cell growth, invasion and migration by targeting LRP6. FEBS J 281:1355–1365
Kahlert UD, Joseph JV, Kruyt FAE (2017) EMT- and MET-related processes in nonepithelial tumors: importance for disease progression, prognosis, and therapeutic opportunities. Mol Oncol 11:860–877
Peixoto P et al (2019) EMT is associated with an epigenetic signature of ECM remodeling genes. Cell Death Dis 10:205
Cook DP, Vanderhyden BC (2020) Context specificity of the EMT transcriptional response. Nat Commun 11:2142
Bernstein BE et al (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326
Chaffer CL et al (2013) Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154:61–74
Pastushenko I, Blanpain C (2019) EMT transition states during tumor progression and metastasis. Trends Cell Biol 29:212–226
Lee K, Gjorevski N, Boghaert E, Radisky DC, Nelson CM (2011) Snail1, Snail2, and E47 promote mammary epithelial branching morphogenesis. EMBO J 30:2662–2674
Welch-Reardon KM et al (2014) Angiogenic sprouting is regulated by endothelial cell expression of Slug. J Cell Sci 127:2017–2028
Stone RC, Pastar I, Ojeh N, Chen V, Liu S, Garzon KI, Tomic-Canic M (2016) Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res 365:495–506
Grande MT et al (2015) Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med 21:989–997
Lovisa S et al (2015) Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med 21:998–1009
Sheng L, Zhuang S (2020) New insights into the role and mechanism of partial epithelial-mesenchymal transition in kidney fibrosis. Front Physiol 11:569322
Puram SV et al (2017) Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171:1611–1624
Armstrong AJ et al (2011) Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 9:997–1007
Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F (2011) Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer 105:1338–1341
Polioudaki H, Agelaki S, Chiotaki R, Politaki E, Mavroudis D, Matikas A, Georgoulias V, Theodoropoulos PA (2015) Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC Cancer 15:399
Wu S et al (2015) Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS ONE 10:e0123976
Baccelli I et al (2013) Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 31:539–544
Boral D et al (2017) Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat Commun 8:196
Liu X et al (2019) Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci Adv 5:eaav4275
Ou H et al (2018) Circulating tumor cell phenotype indicates poor survival and recurrence after surgery for hepatocellular carcinoma. Dig Dis Sci 63:2373–2380
Mani SA et al (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715
Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A (2008) Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE 3:e2888
Celia-Terrassa T et al (2012) Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest 122:1849–1868
Jolly MK, Huang B, Lu M, Mani SA, Levine H, Ben-Jacob E (2014) Towards elucidating the connection between epithelial-mesenchymal transitions and stemness. J R Soc Interface 11:20140962
Jolly MK, Ware KE, Gilja S, Somarelli JA, Levine H (2017) EMT and MET: necessary or permissive for metastasis? Mol Oncol 11:755–769
Pastushenko I et al (2018) Identification of the tumour transition states occurring during EMT. Nature 556:463–468
Fischer KR et al (2015) Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527:472–476
Ren J, Chen Y, Song H, Chen L, Wang R (2013) Inhibition of ZEB1 reverses EMT and chemoresistance in docetaxel-resistant human lung adenocarcinoma cell line. J Cell Biochem 114:1395–1403
Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE (2007) Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol 14:3629–3637
Zheng X et al (2015) Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527:525–530
Burger GA, Danen EHJ, Beltman JB (2017) Deciphering epithelial-mesenchymal transition regulatory networks in cancer through computational approaches. Front Oncol 7:162
Zañudo JG, Guinn MT, Farquhar K, Szenk M, Steinway SN, Balázsi G, Albert R (2019) Towards control of cellular decision-making networks in the epithelial-to-mesenchymal transition. Phys biol 16(3):031002
Dong J et al (2018) Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol 19:31
Lourenco AR et al (2020) Differential contributions of pre- and post-EMT tumor cells in breast cancer metastasis. Cancer Res 80:163–169
McFaline-Figueroa JL, Hill AJ, Qiu X, Jackson D, Shendure J, Trapnell C (2019) A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat Genet 51:1389–1398
Yaswen P (2014) Reinforcing targeted therapeutics with phenotypic stability factors. Cell Cycle 13:3818–3822
Hong T, Watanabe K, Ta CH, Villarreal-Ponce A, Nie Q, Dai X (2015) An Ovol2-Zeb1 mutual inhibitory circuit governs bidirectional and multi-step transition between epithelial and mesenchymal states. PLoS Comput Biol 11:e1004569
Jia D, Jolly MK, Boareto M, Parsana P, Mooney SM, Pienta KJ, Levine H, Ben-Jacob E (2015) OVOL guides the epithelial-hybrid-mesenchymal transition. Oncotarget 6:15436–15448
Watanabe K, Villarreal-Ponce A, Sun P, Salmans ML, Fallahi M, Andersen B, Dai X (2014) Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev Cell 29:59–74
Wu RS et al (2017) OVOL2 antagonizes TGF-beta signaling to regulate epithelial to mesenchymal transition during mammary tumor metastasis. Oncotarget 8:39401–39416
Ren D et al (2014) Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res 358:763–778
Silveira DA, Gupta S, Mombach JCM (2020) Systems biology approach suggests new miRNAs as phenotypic stability factors in the epithelial-mesenchymal transition. J R Soc Interface 17:20200693
Bocci F et al (2019) NRF2 activates a partial epithelial-mesenchymal transition and is maximally present in a hybrid epithelial/mesenchymal phenotype. Integr Biol (Camb) 11:251–263
Subbalakshmi AR, Kundnani D, Biswas K, Ghosh A, Hanash SM, Tripathi SC, Jolly MK (2020) NFATc acts as a non-canonical phenotypic stability factor for a hybrid epithelial/mesenchymal phenotype. Front Oncol 10:553342
Jolly MK, Tripathi SC, Somarelli JA, Hanash SM, Levine H (2017) Epithelial/mesenchymal plasticity: how have quantitative mathematical models helped improve our understanding? Mol Oncol 11:739–754
Hovland AS, Rothstein M, Simoes-Costa M (2020) Network architecture and regulatory logic in neural crest development. Wiley Interdiscip Rev Syst Biol Med 12:e1468
Saunders LR, McClay DR (2014) Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development 141:1503–1513
Herranz H, Cohen SM (2010) MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev 24:1339–1344
Chalancon G, Ravarani CN, Balaji S, Martinez-Arias A, Aravind L, Jothi R, Babu MM (2012) Interplay between gene expression noise and regulatory network architecture. Trends Genet 28:221–232
Lee TI et al (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799–804
Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298:824–827
Ebert MS, Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149:515–524
Re A, Cora D, Taverna D, Caselle M (2009) Genome-wide survey of microRNA-transcription factor feed-forward regulatory circuits in human. Mol Biosyst 5:854–867
Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D (2013) MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Commun 4:2364
Schmiedel JM, Klemm SL, Zheng Y, Sahay A, Bluthgen N, Marks DS, van Oudenaarden A (2015) Gene expression. MicroRNA control of protein expression noise. Science 348:128–132
Cursons J et al (2018) Combinatorial targeting by microRNAs co-ordinates post-transcriptional control of EMT. Cell Syst 7:77–91
Zare H, Khodursky A, Sartorelli V (2014) An evolutionarily biased distribution of miRNA sites toward regulatory genes with high promoter-driven intrinsic transcriptional noise. BMC Evol Biol 14:74
Tripathi S, Xing J, Levine H, Jolly MK (2021) Mathematical modeling of plasticity and heterogeneity in EMT. Methods Mol Biol 2179:385–413
Acknowledgements
The authors would like to acknowledge the critical reading and advice of Prof Greg Goodall, SA Pathology and the University of South Australia, in the preparation of this manuscript. This work was supported by funding from the Australian Research Council (FT190100544, DP190103333) and the Worldwide Cancer Research Foundation (WCR-19-0300).
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. ARC (FT190100544, DP190103333), Worldwide Cancer Research Foundation (WCR-19-0300).
Author information
Authors and Affiliations
Contributions
All three listed authors have contributed to the research and writing of this article.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no competing interests.
Ethics approval
Not required.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Migault, M., Sapkota, S. & Bracken, C.P. Transcriptional and post-transcriptional control of epithelial-mesenchymal plasticity: why so many regulators?. Cell. Mol. Life Sci. 79, 182 (2022). https://doi.org/10.1007/s00018-022-04199-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04199-0