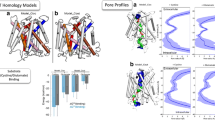
Abstract
Human monocarboxylate transporters (hMCTs/SLC16As) mediate the transport of monocarboxylic compounds across plasma membranes. Among the hMCTs, hMCT1 and hMCT4 are expressed in various tissues, and transport substrates involved in energy metabolism. Both transporters mediate l-lactate transport, but, although hMCT1 also transports l-5-oxoproline (l-OPro), this compound is minimally transported by hMCT4. Thus, we were interested in the molecular mechanism responsible for the difference in substrate specificity between hMCT1 and hMCT4. Therefore, we generated 3D structure models of hMCT1 and hMCT4 to identify amino acid residues involved in the substrate specificity of these transporters. We found that the substrate specificity of hMCT1 was regulated by residues involved in turnover number (M69) and substrate affinity (F367), and these residues were responsible for recognizing (directly or indirectly) the –NH– moiety of l-OPro. Furthermore, our homology model of hMCT1 predicted that M69 and F367 participate in hydrophobic interactions with another region of hMCT1, emphasizing its potentially important role in the binding and translocation cycle of l-OPro. Mutagenesis experiments supported this model, showing that efficient l-OPro transport required a hydrophobic, long linear structure at position 69 and a hydrophobic, γ-branched structure at position 367. Our work demonstrated that the amino acid residues, M69 and F367, are key molecular elements for the transport of l-OPro by hMCT1. These two residues may be involved in substrate recognition and/or substrate-induced conformational changes.






Similar content being viewed by others
Abbreviations
- hMCTs:
-
Human monocarboxylate transporters
- MFS:
-
Major facilitator superfamily
- l-OPro:
-
l-5-oxoproline
- TM:
-
Transmembrane domain
- HMM:
-
Hidden Markov model
- CPC:
-
Cyclopentanecarboxylate
- l-OCPC:
-
(R)-3-Oxocyclopentanecarboxylate
References
Fisel P, Schaeffeler E, Schwab M (2018) Clinical and functional relevance of the monocarboxylate transporter family in disease pathophysiology and drug therapy. Clin Transl Sci 11:352–364. https://doi.org/10.1111/cts.12551
Carneiro L, Pellerin L (2015) Monocarboxylate transporters: new players in body weight regulation. Obes Rev 16:55–66. https://doi.org/10.1111/obr.12256
Pinheiro C, Longatto-Filho A, Azevedo-Silva J et al (2012) Role of monocarboxylate transporters in human cancers: state of the art. J Bioenerg Biomembr 44:127–139. https://doi.org/10.1007/s10863-012-9428-1
Sasaki S, Kobayashi M, Futagi Y et al (2013) Crucial residue involved in L-lactate recognition by human monocarboxylate transporter 4 (hMCT4). PLoS One 8:e67690. https://doi.org/10.1371/journal.pone.0067690
Futagi Y, Sasaki S, Kobayashi M et al (2017) The flexible cytoplasmic loop 3 contributes to the substrate affinity of human monocarboxylate transporters. Biochim Biophys Acta Biomembr 1859:1790–1795. https://doi.org/10.1016/j.bbamem.2017.05.014
Polanski R, Hodgkinson CL, Fusi A et al (2014) Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin Cancer Res 20:926–937. https://doi.org/10.1158/1078-0432.CCR-13-2270
Gurrapu S, Jonnalagadda SK, Alam MA et al (2016) Coumarin carboxylic acids as monocarboxylate transporter 1 inhibitors: in vitro and in vivo studies as potential anticancer agents. Bioorganic Med Chem Lett 26:3282–3286. https://doi.org/10.1016/j.bmcl.2016.05.054
Quanz M, Bender E, Kopitz C et al (2018) Preclinical efficacy of the novel monocarboxylate transporter 1 inhibitor BAY-8002 and associated markers of resistance. Mol Cancer Ther 17:2285–2296. https://doi.org/10.1158/1535-7163.MCT-17-1253
Ovens MJ, Davies AJ, Wilson MC et al (2010) AR-C155858 is a potent inhibitor of monocarboxylate transporters MCT1 and MCT2 that binds to an intracellular site involving transmembrane helices 7–10. Biochem J 425:523–530. https://doi.org/10.1042/BJ20091515
Wang H, Yang C, Doherty JR et al (2014) Synthesis and structure-activity relationships of pteridine dione and trione monocarboxylate transporter 1 inhibitors. J Med Chem 57:7317–7324. https://doi.org/10.1021/jm500640x
Sasaki S, Futagi Y, Kobayashi M et al (2015) Functional characterization of 5-oxoproline transport via SLC16A1/MCT1. J Biol Chem 290:2303–2311. https://doi.org/10.1074/jbc.M114.581892
Bachhawat AK, Yadav S (2018) The glutathione cycle: glutathione metabolism beyond the γ-glutamyl cycle. IUBMB Life 70:585–592. https://doi.org/10.1002/iub.1756
Kumar A, Bachhawat AK (2012) Pyroglutamic acid: throwing light on a lightly studied metabolite. Curr Sci 102:288–297. https://www.jstor.org/stable/24083854
Chou J, Liu R, Yu J et al (2018) Fasting serum α-hydroxybutyrate and pyroglutamic acid as important metabolites for detecting isolated post-challenge diabetes based on organic acid profiles. J Chromatogr B Anal Technol Biomed Life Sci 1100–1101:6–16. https://doi.org/10.1016/j.jchromb.2018.09.004
Qi S, Xu D, Li Q et al (2017) Metabonomics screening of serum identifies pyroglutamate as a diagnostic biomarker for nonalcoholic steatohepatitis. Clin Chim Acta 473:89–95. https://doi.org/10.1016/j.cca.2017.08.022
Yu B, Zheng Y, Nettleton JA et al (2014) Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol 9:1410–1417. https://doi.org/10.2215/CJN.11971113
Thiesen L, Kehler J, Clausen RP et al (2015) In vitro and in vivo evidence for active brain uptake of the GHB analog HOCPCA by the monocarboxylate transporter subtype 1. J Pharmacol Exp Ther 354:166–174. https://doi.org/10.1124/jpet.115.224543
Futagi Y, Kobayashi M, Narumi K et al (2018) Identification of a selective inhibitor of human monocarboxylate transporter 4. Biochem Biophys Res Commun 495:427–432. https://doi.org/10.1016/j.bbrc.2017.10.025
Galić S, Schneider H-PP, Bröer A et al (2003) The loop between helix 4 and helix 5 in the monocarboxylate transporter (MCT1) is important for substrate selection and protein stability. Biochem J 376:413–422. https://doi.org/10.1042/BJ20030799
Garcia CK, Goldstein JL, Pathak RK et al (1994) Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: implications for the Cori cycle. Cell 76:865–873. https://doi.org/10.1016/0092-8674(94)90361-1
Rahman B, Schneider HP, Bröer A et al (1999) Helix 8 and helix 10 are involved in substrate recognition in the rat monocarboxylate transporter MCT1. Biochemistry 38:11577–11584. https://doi.org/10.1021/bi990973f
Deng D, Sun P, Yan C et al (2015) Molecular basis of ligand recognition and transport by glucose transporters. Nature 526:391–396. https://doi.org/10.1038/nature14655
Zhao Y, Heng J, Zhao Y et al (2015) Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res 25:1060–1073. https://doi.org/10.1038/cr.2015.94
Parker JL, Newstead S (2014) Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507:68–72. https://doi.org/10.1038/nature13116
Sun J, Bankston JR, Payandeh J et al (2014) Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507:73–77. https://doi.org/10.1038/nature13074
Pedersen BP, Kumar H, Waight AB et al (2013) Crystal structure of a eukaryotic phosphate transporter. Nature 496:533–536. https://doi.org/10.1038/nature12042
Yan H, Huang W, Yan C et al (2013) Structure and mechanism of a nitrate transporter. Cell Rep 3:716–723. https://doi.org/10.1016/j.celrep.2013.03.007
Sun L, Zeng X, Yan C et al (2012) Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 490:361–366. https://doi.org/10.1038/nature11524
Nancolas B, Sessions RB, Halestrap AP (2015) Identification of key binding site residues of MCT1 for AR-C155858 reveals the molecular basis of its isoform selectivity. Biochem J 466:177–188. https://doi.org/10.1042/BJ20141223
Johannes J, Braun D, Kinne A et al (2016) Few amino acid exchanges expand the substrate spectrum of monocarboxylate transporter 10. Mol Endocrinol 30:796–808. https://doi.org/10.1210/me.2016-1037
Edwards N, Anderson CMH, Conlon NJ et al (2018) Resculpting the binding pocket of APC superfamily LeuT-fold amino acid transporters. Cell Mol Life Sci 75:921–938. https://doi.org/10.1007/s00018-017-2677-8
Krypotou E, Kosti V, Amillis S et al (2012) Modeling, substrate docking, and mutational analysis identify residues essential for the function and specificity of a eukaryotic purine-cytosine NCS1 transporter. J Biol Chem 287:36792–36803. https://doi.org/10.1074/jbc.M112.400382
Monné M, Miniero DV, Daddabbo L et al (2012) Substrate specificity of the two mitochondrial ornithine carriers can be swapped by single mutation in substrate binding site. J Biol Chem 287:7925–7934. https://doi.org/10.1074/jbc.M111.324855
Gui C, Hagenbuch B (2008) Amino acid residues in transmembrane domain 10 of organic anion transporting polypeptide 1B3 are critical for cholecystokinin octapeptide transport. Biochemistry 47:9090–9097. https://doi.org/10.1021/bi8008455
Sasaki S, Futagi Y, Ideno M et al (2016) Effect of diclofenac on SLC16A3/MCT4 by the Caco-2 cell line. Drug Metab Pharmacokinet 31:218–223. https://doi.org/10.1016/j.dmpk.2016.03.004
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. Embnet News 4:1–4
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015-053
Remmert M, Biegert A, Hauser A, Söding J (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9:173–175. https://doi.org/10.1038/nmeth.1818
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. https://doi.org/10.1006/jmbi.1999.3091
Söding J (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. https://doi.org/10.1093/bioinformatics/bti125
Jefferys BR, Kelley LA, Sternberg MJE (2010) Protein folding requires crowd control in a simulated cell. J Mol Biol 397:1329–1338. https://doi.org/10.1016/j.jmb.2010.01.074
Xie W, Sahinidis NV (2006) Residue-rotamer-reduction algorithm for the protein side-chain conformation problem. Bioinformatics 22:188–194. https://doi.org/10.1093/bioinformatics/bti763
Huang Y, Lemieux MJ, Song J et al (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301:616–620. https://doi.org/10.1126/science.1087619
Iancu CV, Zamoon J, Woo SB et al (2013) Crystal structure of a glucose/H + symporter and its mechanism of action. Proc Natl Acad Sci 110:17862–17867. https://doi.org/10.1073/pnas.1311485110
Jiang D, Zhao Y, Wang X et al (2013) Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc Natl Acad Sci 110:14664–14669. https://doi.org/10.1073/pnas.1308127110
Bhattacharya D, Nowotny J, Cao R, Cheng J (2016) 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res 44:W406–W409. https://doi.org/10.1093/nar/gkw336
Lovell SC, Davis IW, Arendall WB III et al (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins: structure. Funct Genet 50:437–450
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:327–331. https://doi.org/10.1093/nar/gkh454
Marchler-Bauer A, Bo Y, Han L et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Menke M, Berger B, Cowen L (2008) Matt: local flexibility aids protein multiple structure alignment. PLoS Comput Biol 4:0088–0099. https://doi.org/10.1371/journal.pcbi.0040010
Kim S, Chen J, Cheng T et al (2019) PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 47:D1102–D1109. https://doi.org/10.1093/nar/gky1033
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Shapovalov MV, Dunbrack RL (2011) A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19:844–858. https://doi.org/10.1016/j.str.2011.03.019
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247–260. https://doi.org/10.1016/j.jmgm.2005.12.005
Trott oleg O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc
Vivian D, Polli JE (2014) Mechanistic interpretation of conventional Michaelis-Menten parameters in a transporter system. Eur J Pharm Sci 64:44–52. https://doi.org/10.1016/j.ejps.2014.08.007
Groeneweg S, Friesema ECH, Kersseboom S et al (2014) The role of Arg445 and Asp498 in the human thyroid hormone transporter MCT8. Endocrinology 155:618–626. https://doi.org/10.1210/en.2013-1521
Quistgaard EM, Löw C, Guettou F, Nordlund P (2016) Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat Rev Mol Cell Biol 17:123–132. https://doi.org/10.1038/nrm.2015.25
Perland E, Fredriksson R (2017) Classification systems of secondary active transporters. Trends Pharmacol Sci 38:305–315. https://doi.org/10.1016/j.tips.2016.11.008
Stäubli A, Capatina N, Fuhrer Y et al (2017) Abnormal creatine transport of mutations in monocarboxylate transporter 12 (MCT12) found in patients with age-related cataract can be partially rescued by exogenous chaperone CD147. Hum Mol Genet 26:4203–4214. https://doi.org/10.1093/hmg/ddx310
Manoharan C, Wilson MC, Sessions RB, Halestrap AP (2006) The role of charged residues in the transmembrane helices of monocarboxylate transporter 1 and its ancillary protein basigin in determining plasma membrane expression and catalytic activity. Mol Membr Biol 23:486–498. https://doi.org/10.1080/09687860600841967
Islam MS, Namba N, Ohata Y et al (2018) Functional analysis of monocarboxylate transporter 8 mutations in Japanese Allan-Herndon-Dudley syndrome patients. Endocr J 2018:1–11. https://doi.org/10.1507/endocrj.EJ18-0251
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS; KAKENHI Grant Number JP17J00013). We would like to thank Editage (http://www.editage.jp) for English language editing.
Author information
Authors and Affiliations
Contributions
YF designed and performed the experiments, analyzed the results, and wrote the first draft of the manuscript. MK, KN, AF, and KI contributed to the writing of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Futagi, Y., Kobayashi, M., Narumi, K. et al. Homology modeling and site-directed mutagenesis identify amino acid residues underlying the substrate selection mechanism of human monocarboxylate transporters 1 (hMCT1) and 4 (hMCT4). Cell. Mol. Life Sci. 76, 4905–4921 (2019). https://doi.org/10.1007/s00018-019-03151-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03151-z