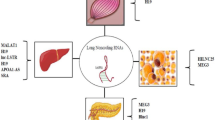
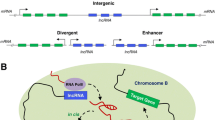
Abstract
Type 2 diabetes is a complex metabolic disorder characterized by insulin resistance and pancreatic β-cell dysfunction. Deregulated glucose and lipid metabolism are the primary underlying manifestations associated with this disease and its complications. Long non-coding RNAs (lncRNAs) are a novel class of functional RNAs that regulate a variety of biological processes by a diverse interplay of mechanisms including recruitment of epigenetic modifiers, transcriptional and post-transcriptional regulation, control of mRNA decay, and sequestration of transcription factors. Although the underlying causes that define the diabetic phenotype are extremely intricate, most of the studies in the last decades were mostly centered on protein-coding genes. However, current opinion in the recent past has authenticated the contributions of diverse lncRNAs as critical regulatory players during the manifestation of diabetes. The current review will highlight the importance of lncRNAs in regulating cellular processes that govern metabolic homeostasis in key metabolic tissues. A more in-depth understanding of lncRNAs may enable their exploitation as biomarkers or for therapeutic applications during diabetes and its associated complications.




Similar content being viewed by others
References
Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74
Mattick JS (2004) RNA regulation: a new genetics? Nat Rev Genet 5:316–323
Bánfai B, Jia H, Khatun J et al (2012) Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22:1646–1657
Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147:789–802
Tupy JL, Bailey AM, Dailey G et al (2005) Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci USA 102:5495–5500
Kondo T, Plaza S, Zanet J et al (2010) Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329:336–339
Cabili MN, Trapnell C, Goff L et al (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25:1915–1927
Carninci P, Kasukawa T, Katayama S et al (2005) The transcriptional landscape of the mammalian genome. Science 309:1559–1563
Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462:799–802
Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M (2008) A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32:685–695
Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH (2007) Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316:1345–1348
Bernstein HD, Zopf D, Freymann DM, Walter P (1993) Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci USA 90:5229–5233
Clemson CM, McNeil JA, Willard HF, Lawrence JB (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132:259–275
Derrien T, Johnson R, Bussotti G et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789
Brosius J (2005) Waste not, want not-transcript excess in multicellular eukaryotes. Trends Genet 21:287–288
Djebali S, Davis CA, Merkel A et al (2012) Landscape of transcription in human cells. Nature 489:101–108
Fatica A, Bozzoni I (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15:7–21
Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21:1253–1261
Yu Y, Fuscoe JC, Zhao C et al (2014) A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5:3230
Kaushik K, Leonard VE, Kv S et al (2013) Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS One 8:e83616
Guttman M, Amit I, Garber M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227
Huarte M, Guttman M, Feldser D et al (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142:409–419
Prensner JR, Iyer MK, Balbin OA et al (2011) Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29:742–749
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10:924–933
Katayama S, Tomaru Y, Kasukawa T et al (2005) Antisense transcription in the mammalian transcriptome. Science 309:1564–1566
Duvel K, Yecies JL, Menon S et al (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39:171–183
Mousavi K, Zare H, Dell’orso S et al (2013) eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51:606–617
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914
Wang KC, Yang YW, Liu B et al (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472:120–124
Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB (2012) Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One 7:e47998
Li D, Feng J, Wu T et al (2013) Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol 182:64–70
Brown CJ, Ballabio A, Rupert JL et al (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38–44
Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E (2006) RNA-mediated response to heat shock in mammalian cells. Nature 440:556–560
Beltran M, Puig I, Pena C et al (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22:756–769
van de Vondervoort II, Gordebeke PM, Khoshab N et al (2013) Long non-coding RNAs in neurodevelopmental disorders. Front Mol Neurosci 6:53
Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS (2016) LncRNAs and cancer. Oncol Lett 12:1233–1239
Li J, Xuan Z, Liu C (2013) Long non-coding RNAs and complex human diseases. Int J Mol Sci 14:18790–18808
Matouk IJ, DeGroot N, Mezan S et al (2007) The H19 non-coding RNA is essential for human tumor growth. PLoS One 2:e845
Berteaux N, Lottin S, Monte D et al (2005) H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 280:29625–29636
Barsyte-Lovejoy D, Lau SK, Boutros PC et al (2006) The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 66:5330–5337
Yang F, Bi J, Xue X et al (2012) Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 279:3159–3165
Li H, Yu B, Li J et al (2014) Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5:2318–2329
Tu ZQ, Li RJ, Mei JZ, Li XH (2014) Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol 7:4303–4309
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH (2015) Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol 36:4851–4859
Liu Y, Zhang M, Liang L, Li J, Chen YX (2015) Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol 8:11480–11484
Kumarswamy R, Bauters C, Volkmann I et al (2014) Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 114:1569–1575
Kornfeld JW, Bruning JC (2014) Regulation of metabolism by long, non-coding RNAs. Front Genet 5:57
Knoll M, Lodish HF, Sun L (2015) Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol 11:151–160
Hanson RL, Craig DW, Millis MP et al (2007) Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 56:975–983
Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG (2010) The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet 42:68–71
Fadista J, Vikman P, Laakso EO et al (2014) Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA 111:13924–13929
Motterle A, Sanchez-Parra C, Regazzi R (2016) Role of long non-coding RNAs in the determination of beta-cell identity. Diabetes Obes Metab 18(Suppl 1):41–50
Eliasson L, Esguerra JL (2014) Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol (Oxf) 211:273–284
Ding GL, Wang FF, Shu J et al (2012) Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 61:1133–1142
Ku GM, Kim H, Vaughn IW et al (2012) Research resource: RNA-Seq reveals unique features of the pancreatic beta-cell transcriptome. Mol Endocrinol 26:1783–1792
Moran I, Akerman I, van de Bunt M et al (2012) Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 16:435–448
Yin DD, Zhang EB, You LH et al (2015) Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic beta cells. Cell Physiol Biochem 35:1892–1904
You L, Wang N, Yin D et al (2016) Downregulation of long noncoding RNA Meg3 affects insulin synthesis and secretion in mouse pancreatic beta cells. J Cell Physiol 231:852–862
Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L (2016) betalinc1 encodes a long noncoding RNA that regulates islet beta-cell formation and function. Genes Dev 30:502–507
Akerman I, Tu Z, Beucher A et al (2017) Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metab 25:400–411
Li P, Ruan X, Yang L et al (2015) A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 21:455–467
Halley P, Kadakkuzha BM, Faghihi MA et al (2014) Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep 6:222–230
Hu YW, Yang JY, Ma X et al (2014) A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J Lipid Res 55:681–697
Cui M, Xiao Z, Wang Y et al (2015) Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res 75:846–857
Zhu X, Wu YB, Zhou J, Kang DM (2016) Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem Biophys Res Commun 469:319–325
Ruan X, Li P, Cangelosi A, Yang L, Cao H (2016) A long non-coding RNA, lncLGR, regulates hepatic glucokinase expression and glycogen storage during fasting. Cell Rep 14:1867–1875
Li D, Cheng M, Niu Y et al (2017) Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int J Biol Sci 13:349–357
Goyal N, Sivadas A, Shamsudheen K et al (2017) RNA sequencing of db/db mice liver identifies lncRNA H19 as a key regulator of gluconeogenesis and hepatic glucose output. Sci Rep 7:8312
Pope C, Mishra S, Russell J, Zhou Q, Zhong X-B (2017) Targeting H19, an imprinted long non-coding RNA, in hepatic functions and liver diseases. Diseases 5:11
Gao Y, Wu F, Zhou J et al (2014) The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res 42:13799–13811
Kallen AN, Zhou XB, Xu J et al (2013) The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 52:101–112
Dey BK, Pfeifer K, Dutta A (2014) The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 28:491–501
Zhao XY, Lin JD (2015) Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci 40:586–596
Wang L, Zhao Y, Bao X et al (2015) LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res 25:335–350
Cesana M, Cacchiarelli D, Legnini I et al (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369
Divoux A, Karastergiou K, Xie H et al (2014) Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity (Silver Spring) 22:1781–1785
Zhao XY, Li S, Wang GX, Yu Q, Lin JD (2014) A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 55:372–382
Wei S, Du M, Jiang Z, Hausman GJ, Zhang L, Dodson MV (2016) Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity. Cell Mol Life Sci 73:2079–2087
Xu B, Gerin I, Miao H et al (2010) Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One 5:e14199
Xiao T, Liu L, Li H et al (2015) Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha. Stem Cell Reports 5:856–865
Wang F, Tong Q (2008) Transcription factor PU.1 is expressed in white adipose and inhibits adipocyte differentiation. Am J Physiol Cell Physiol 295:C213–C220
Liu J, Yao J, Li X et al (2014) Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 5:e1506
Yan B, Tao Z-F, Li X-M, Zhang H, Yao J, Jiang Q (2014) Aberrant expression of long noncoding RNAs in early diabetic retinopathy aberrant expression of lncRNAs in early DR. Investig Ophthalmol Vis Sci 55:941–951
Qiu G-Z, Tian W, Fu H-T, Li C-P, Liu B (2016) Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Biophys Res Commun 471:135–141
Thomas AA, Feng B, Chakrabarti S (2017) ANRIL: a regulator of VEGF in diabetic retinopathy role of ANRIL in diabetic retinopathy. Investig Ophthalmol Vis Sci 58:470–480
Siddiqui A, Hussain S, Azam A et al (2017) ANRIL polymorphism rs1333049, a novel genetic predictor for diabetic retinopathy complication. Meta Gene 14:33–37
Fox CS, Larson MG, Leip EP, Meigs JB, Wilson PW, Levy D (2005) Glycemic status and development of kidney disease. Diabetes Care 28:2436–2440
Alvarez ML, DiStefano JK (2011) Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One 6:e18671
Alvarez ML, Khosroheidari M, Eddy E, Kiefer J (2013) Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS One 8:e77468
Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X (2015) Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun 468:726–732
Kato M, Wang M, Chen Z et al (2016) An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 7:12864
Yi H, Peng R, L-y Zhang et al (2017) LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis 8:e2583
Duan L-J, Ding M, Hou L-J, Cui Y-T, Li C-J, Yu D-M (2017) Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem Biophys Res Commun 484:598–604
Aneja A, Tang WW, Bansilal S, Garcia MJ, Farkouh ME (2008) Diabetic cardiomyopathy: insights into pathogenesis, diagnostic challenges, and therapeutic options. Am J Med 121:748–757
Zhang M, Gu H, Chen J, Zhou X (2016) Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int J Cardiol 202:753–755
Zhang M, Gu H, Xu W, Zhou X (2016) Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int J Cardiol 203:214–216
Shi Y, Wang Y, Luan W et al (2014) Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One 9:e86295
Li X, Wang H, Yao B, Xu W, Chen J, Zhou X (2016) lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci Rep 6:36340
Peng H, Zou L, Xie J et al (2017) lncRNA NONRATT021972 siRNA decreases diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root ganglia. Mol Neurobiol 54:511–523
Wang S, Xu H, Zou L et al (2016) LncRNA uc. 48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal 12:139–148
Zgheib C, Hodges MM, Hu J, Liechty KW, Xu J (2017) Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PLoS One 12:e0177453
Bolha L, Ravnik-Glavač M, Glavač D (2017) Long noncoding RNAs as biomarkers in cancer. Dis Markers 2017:7243968
Shi T, Gao G, Cao Y (2016) Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers 2016:9085195
Bussemakers MJ, van Bokhoven A, Verhaegh GW et al (1999) DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 59:5975–5979
Zhou X, Yin C, Dang Y, Ye F, Zhang G (2015) Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep 5:11516
Panzitt K, Tschernatsch MM, Guelly C et al (2007) Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132:330–342
Wang X-S, Zhang Z, Wang H-C et al (2006) Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res 12:4851–4858
Deng R, Liu B, Wang Y et al (2016) High expression of the newly found long noncoding RNA Z38 promotes cell proliferation and oncogenic activity in breast cancer. J Cancer 7:576–586
Tang J, Zhuo H, Zhang X et al (2014) A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis 5:e1549
Hanna N, Ohana P, Konikoff FM et al (2012) Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther 19:374–381
Amit D, Hochberg A (2010) Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J Transl Med 8:134
Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T (2016) Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther 161:67–78
Mizrahi A, Czerniak A, Levy T et al (2009) Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med 7:69
Acknowledgements
This work was supported by funding from the Council of Scientific and Industrial Research (CSIR), New Delhi, India (BSC0123). NG and DK acknowledge CSIR, New Delhi, India for their fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None to declare.
Rights and permissions
About this article
Cite this article
Goyal, N., Kesharwani, D. & Datta, M. Lnc-ing non-coding RNAs with metabolism and diabetes: roles of lncRNAs. Cell. Mol. Life Sci. 75, 1827–1837 (2018). https://doi.org/10.1007/s00018-018-2760-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2760-9