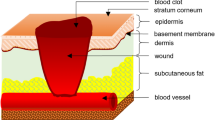
Abstract
Cell-generated forces drive an array of biological processes ranging from wound healing to tumor metastasis. Whereas experimental techniques such as traction force microscopy are capable of quantifying traction forces in multidimensional systems, the physical mechanisms by which these forces induce changes in tissue form remain to be elucidated. Understanding these mechanisms will ultimately require techniques that are capable of quantifying traction forces with high precision and accuracy in vivo or in systems that recapitulate in vivo conditions, such as microfabricated tissues and engineered substrata. To that end, here we review the fundamentals of traction forces, their quantification, and the use of microfabricated tissues designed to study these forces during cell migration and tissue morphogenesis. We emphasize the differences between traction forces in two- and three-dimensional systems, and highlight recently developed techniques for quantifying traction forces.



Similar content being viewed by others
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- AFM:
-
Atomic force microscopy
- ECM:
-
Extracellular matrix
- FRET:
-
Fluorescence resonance energy transfer
- MDCK:
-
Madin–Darby canine kidney
- PA:
-
Polyacrylamide
- PEG:
-
Polyethylene glycol
- TFM:
-
Traction force microscopy
References
Maniotis AJ, Chen CS, Ingber DE (1997) Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA 94(3):849–854. doi:10.1073/pnas.94.3.849
Thery M, Jimenez-Dalmaroni A, Racine V, Bornens M, Julicher F (2007) Experimental and theoretical study of mitotic spindle orientation. Nature 447(7143):493–496. doi:10.1038/nature05786
Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE (1997) Geometric control of cell life and death. Science 276(5317):1425–1428. doi:10.1126/science.276.5317.1425
Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126(4):677–689. doi:10.1016/j.cell.2006.06.044
Desprat N, Supatto W, Pouille P-A, Beaurepaire E, Farge E (2008) Tissue deformation modulates twist expression to determine anterior midgut differentiation in Drosophila embryos. Dev Cell 15(3):470–477. doi:10.1016/j.devcel.2008.07.009
Farge E (2003) Mechanical induction of twist in the Drosophila foregut/stomodeal primordium. Curr Biol 13(16):1365–1377. doi:10.1016/S0960-9822(03)00576-1
Hinz B, Mastrangelo D, Iselin CE, Chaponnier C, Gabbiani G (2001) Mechanical tension controls granulation tissue contractile activity and myofibroblast differentiation. Am J Pathol 159(3):1009–1020. doi:10.1016/S0002-9440(10)61776-2
Munevar S, Wang Y, Dembo M (2001) Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J 80(4):1744–1757. doi:10.1016/S0006-3495(01)76145-0
Peschetola V, Laurent VM, Duperray A, Michel R, Ambrosi D, Preziosi L, Verdier C (2013) Time-dependent traction force microscopy for cancer cells as a measure of invasiveness. Cytoskeleton. doi:10.1002/cm.21100 (1949-3592 (Electronic))
Koch TM, Münster S, Bonakdar N, Butler JP, Fabry B (2012) 3D Traction forces in cancer cell invasion. PLoS One 7(3):e33476. doi:10.1371/journal.pone.0033476
Reffay M, Parrini MC, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, Silberzan P (2014) Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat Cell Biol 16(3):217–223. doi:10.1038/ncb2917
Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ (2009) Physical forces during collective cell migration. Nat Phys 5(6):426–430. doi:10.1038/nphys1269
Heisenberg C-P, Bellaïche Y (2013) Forces in tissue morphogenesis and patterning. Cell 153(5):948–962. doi:10.1016/j.cell.2013.05.008
Gov NS (2009) Traction forces during collective cell motion. HFSP Journal 3(4):223–227. doi:10.2976/1.3185785
His W (1874) Unsere Körperform und das physiologische Problem ihrer Entstehung Briefe an einen befreundeten Naturforscher/von Wilhelm His. F.C.W. Vogel, Leipzig. doi:10.5962/bhl.title.28860
Paluch EK, Nelson CM, Biais N, Fabry B, Moeller J, Pruitt BL, Wollnik C, Kudryasheva G, Rehfeldt F, Federle W (2015) Mechanotransduction: use the force(s). BMC Biol 13(1):1–14. doi:10.1186/s12915-015-0150-4
Richards RJ (2008) The tragic sense of life: ernst haeckel and the struggle over evolutionary thought. University of Chicago Press, Chicago
Maskarinec SA, Franck C, Tirrell DA, Ravichandran G (2009) Quantifying cellular traction forces in three dimensions. Proc Natl Acad Sci USA 106(52):22108–22113. doi:10.1073/pnas.0904565106
Mok S, Moraes C (2016) Thinking big by thinking small: advances in mechanobiology across the length scales. Integr Biol 8(3):262–266. doi:10.1039/C6IB90008A
Polacheck WJ, Chen CS (2016) Measuring cell-generated forces: a guide to the available tools. Nat Meth 13(5):415–423. doi:10.1038/nmeth.3834
Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, Wilen LA, Xu Y, Dufresne ER (2014) Traction force microscopy in physics and biology. Soft Matter 10(23):4047–4055. doi:10.1039/C4SM00264D
Ribeiro AJS, Denisin AK, Wilson RE, Pruitt BL (2016) For whom the cells pull: hydrogel and micropost devices for measuring traction forces. Methods 94:51–64. doi:10.1016/j.ymeth.2015.08.005
Gupta M, Kocgozlu L, Sarangi BR, Margadant F, Ashraf M, Ladoux B (2015) Chapter 16—micropillar substrates: a tool for studying cell mechanobiology. In: Ewa KP (ed) Methods in cell biology, vol 125. Academic Press, London, pp 289–308. doi:10.1016/bs.mcb.2014.10.009
Nelson CM, Tien J (2006) Microstructured extracellular matrices in tissue engineering and development. Curr Opin Biotechnol 17(5):518–523. doi:10.1016/j.copbio.2006.08.011
Tien J, Nelson CM (2013) Microstructured extracellular matrices in tissue engineering and development: an update. Ann Biomed Eng 42(7):1413–1423. doi:10.1007/s10439-013-0912-5
Zorlutuna P, Annabi N, Camci-Unal G, Nikkhah M, Cha JM, Nichol JW, Manbachi A, Bae H, Chen S, Khademhosseini A (2012) Microfabricated biomaterials for engineering 3D tissues. Adv Mater 24(14):1782–1804. doi:10.1002/adma.201104631
Harris AK, Wild P, Stopak D (1980) Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208(4440):177–179. doi:10.1126/science.6987736
Harris JK (1978) A photoelastic substrate technique for dynamic measurements of forces exerted by moving organisms. J Microsc 114(2):219–228. doi:10.1111/j.1365-2818.1978.tb00132.x
Harris AK (1973) Cell surface movements related to cell locomotion. In: Locomotion of tissue cells, Ciba Foundation Symposium, Amsterdam, Elsevier, pp 3–26. doi:10.1002/9780470719978.ch2
Dembo M, Wang Y-L (1999) Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J 76(4):2307–2316. doi:10.1016/S0006-3495(99)77386-8
Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K (1994) Traction forces generated by locomoting keratocytes. J Cell Biol 127(6):1957–1964. doi:10.1083/jcb.127.6.1957
Oliver T, Jacobson K, Dembo M (1995) Traction forces in locomoting cells. Cell Motil Cytoskeleton 31(3):225–240. doi:10.1002/cm.970310306
Burton K, Taylor DL (1997) Traction forces of cytokinesis measured with optically modified elastic substrata. Nature 385(6615):450–454. doi:10.1038/385450a0
Roy P, Petroll WM, Cavanagh HD, Chuong CJ, Jester JV (1997) An in vitro force measurement assay to study the early mechanical interaction between corneal fibroblasts and collagen matrix. Exp Cell Res 232(1):106–117. doi:10.1006/excr.1997.3511
Brandley BK, Weisz OA, Schnaar RL (1987) Cell attachment and long-term growth on derivatizable polyacrylamide surfaces. J Biol Chem 262(13):6431–6437
Pelham RJ, Wang Y-L (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA 94(25):13661–13665. doi:10.1073/pnas.94.25.13661
Wang Y-L, Pelham RJ Jr (1998) Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods in Enzymology, vol 298. Academic Press, London, pp 489–496. doi:10.1016/S0076-6879(98)98041-7
Beningo KA, Wang Y-L (2002) Flexible substrata for the detection of cellular traction forces. Trends Cell Biol 12(2):79–84. doi:10.1016/S0962-8924(01)02205-X
Sabass B, Gardel ML, Waterman CM, Schwarz US (2008) High resolution traction force microscopy based on experimental and computational advances. Biophys J 94(1):207–220. doi:10.1529/biophysj.107.113670
Lautscham LA, Lin CY, Auernheimer V, Naumann CA, Goldmann WH, Fabry B (2014) Biomembrane-mimicking lipid bilayer system as a mechanically tunable cell substrate. Biomaterials 35(10):3198–3207. doi:10.1016/j.biomaterials.2013.12.091
Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS (2003) Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA 100(4):1484–1489. doi:10.1073/pnas.0235407100
Galbraith CG, Sheetz MP (1997) A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci USA 94(17):9114–9118. doi:10.1073/pnas.94.17.9114
Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B (2001) Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol 3(5):466–472. doi:10.1038/35074532
Rodriguez ML, Graham BT, Pabon LM, Han SJ, Murry CE, Sniadecki NJ (2014) Measuring the contractile forces of human induced pluripotent stem cell-derived cardiomyocytes with arrays of microposts. J Biomech Eng 136(5):051005. doi:10.1115/1.4027145
du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Siberzan P, Ladoux B (2005) Force mapping in epithelial cell migration. Proc Natl Acad Sci USA 102(7):2390–2395. doi:10.1073/pnas.0408482102
Schoen I, Hu W, Klotzsch E, Vogel V (2010) Probing cellular traction forces by micropillar arrays: contribution of substrate warping to pillar deflection. Nano Lett 10(5):1823–1830. doi:10.1021/nl100533c
Hur SS, Zhao Y, Li Y-S, Botvinick E, Chien S (2009) Live cells exert 3-dimensional traction forces on their substrata. Cell Mol Bioeng 2(3):425–436. doi:10.1007/s12195-009-0082-6
del Álamo JC, Meili R, Álvarez-González B, Alonso-Latorre B, Bastounis E, Firtel R, Lasheras JC (2013) Three-dimensional quantification of cellular traction forces and mechanosensing of thin substrata by Fourier traction force microscopy. PLoS One 8(9):e69850. doi:10.1371/journal.pone.0069850
Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS (2013) Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc Natl Acad Sci USA 110(3):881–886. doi:10.1073/pnas.1207997110
Delanoë-Ayari H, Rieu JP, Sano M (2010) 4D traction force microscopy reveals asymmetric cortical forces in migrating dictyostelium cells. Phys Rev Lett 105(24):248103. doi:10.1103/PhysRevLett.105.248103
Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS (2010) Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Meth 7(12):969–971. doi:10.1038/nmeth.1531
Steinwachs J, Metzner C, Skodzek K, Lang N, Thievessen I, Mark C, Munster S, Aifantis KE, Fabry B (2016) Three-dimensional force microscopy of cells in biopolymer networks. Nat Meth 13(2):171–176. doi:10.1038/nmeth.3685
Hall MS, Long R, Feng X, Huang Y, Hui C-Y, Wu M (2013) Toward single cell traction microscopy within 3D collagen matrices. Exp Cell Res 319(16):2396–2408. doi:10.1016/j.yexcr.2013.06.009
Gjorevski N, Piotrowski AS, Varner VD, Nelson CM (2015) Dynamic tensile forces drive collective cell migration through three-dimensional extracellular matrices. Sci Rep 5:11458. doi:10.1038/srep11458
Gjorevski N, Nelson CM (2012) Mapping of mechanical strains and stresses around quiescent engineered three-dimensional epithelial tissues. Biophys J 103(1):152–162. doi:10.1016/j.bpj.2012.05.048
Piotrowski AS, Varner VD, Gjorevski N, Nelson CM (2015) Three-dimensional traction force microscopy of engineered epithelial tissues. In: Nelson CM (ed) Tissue morphogenesis: methods and protocols. Springer, New York, pp 191–206. doi:10.1007/978-1-4939-1164-6_13
Landau LD, Lifshitz EM (1986) Theory of elasticity, vol 7. Course of theoretical physics, 3 edn. Elsevier Butterworth-Heinemann, Institute of Physical Problems, USSR Academy of Sciences, Moscow USSR
Landau LD, Lifshitz EM (1976) Mechanics, vol 1. Course of Theoretical Physics, 3 edn. Elsevier Butterworth-Heinemann, Institute of Physical Problems, USSR Academy of Sciences, Moscow USSR
Butler JP, Tolić-Nørrelykke IM, Fabry B, Fredberg JJ (2002) Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol 282(3):C595–C605. doi:10.1152/ajpcell.00270.2001
Ambrosi D (2006) Cellular Traction as an Inverse Problem. SIAM J Numer Anal 66(6):2049–2060. doi:10.1137/060657121
Stout DA, Bar-Kochba E, Estrada JB, Toyjanova J, Kesari H, Reichner JS, Franck C (2016) Mean deformation metrics for quantifying 3D cell–matrix interactions without requiring information about matrix material properties. Proc Natl Acad Sci USA 113(11):2898–2903. doi:10.1073/pnas.1510935113
Franck C, Hong S, Maskarinec SA, Tirrell DA, Ravichandran G (2007) Three-dimensional full-field measurements of large deformations in soft materials using confocal microscopy and digital volume correlation. Exp Mech 47(3):427–438. doi:10.1007/s11340-007-9037-9
Bar-Kochba E, Toyjanova J, Andrews E, Kim KS, Franck C (2015) A fast iterative digital volume correlation algorithm for large deformations. Exp Mech 55(1):261–274. doi:10.1007/s11340-014-9874-2
Dembo M, Oliver T, Ishihara A, Jacobson K (1996) Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J 70(4):2008–2022. doi:10.1016/S0006-3495(96)79767-9
Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, Safran SA (2002) Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys J 83(3):1380–1394. doi:10.1016/S0006-3495(02)73909-X
Hansen P (1998) Rank-deficient and discrete ill-posed problems Mathematical modeling and computation. Soc Ind Appl Math. doi:10.1137/1.9780898719697
Schwarz US (1853) Soiné JRD (2015) Traction force microscopy on soft elastic substrates: a guide to recent computational advances. Biochim Biophys Acta 11, Part B:3095–3104. doi:10.1016/j.bbamcr.2015.05.028
Plotnikov SV, Sabass B, Schwarz US, Waterman CM (2014) Chapter 20—high-resolution traction force microscopy. In: Jennifer CW, Torsten W (eds) Methods in cell biology, vol 123. Academic Press, London, pp 367–394. doi:10.1016/B978-0-12-420138-5.00020-3
Vaughan RB, Trinkaus JP (1966) Movements of epithelial cell sheets in vitro. J Cell Sci 1(4):407–413
Siedlik MJ, Varner VD, Nelson CM (2016) Pushing, pulling, and squeezing our way to understanding mechanotransduction. Methods. doi:10.1016/j.ymeth.2015.08.019
Borghi N, Farge E, Lavelle C (2016) Experimental approaches in mechanotransduction: from molecules to pathology. Methods 94:1–3. doi:10.1016/j.ymeth.2016.01.007
Miller CJ, Davidson LA (2013) The interplay between cell signalling and mechanics in developmental processes. Nat Rev Genet 14(10):733–744. doi:10.1038/nrg3513
Molino D, Quignard S, Gruget C, Pincet F, Chen Y, Piel M, Fattaccioli J (2016) On-chip quantitative measurement of mechanical stresses during cell migration with emulsion droplets. Sci Rep 6:29113. doi:10.1038/srep29113
Mann C, Leckband D (2010) Measuring traction forces in long-term cell cultures. Cell Mol Bioeng 3(1):40–49. doi:10.1007/s12195-010-0108-0
Jannat RA, Dembo M, Hammer DA (2011) Traction forces of neutrophils migrating on compliant substrates. Biophys J 101(3):575–584. doi:10.1016/j.bpj.2011.05.040
Ricart BG, Yang MT, Hunter CA, Chen CS, Hammer DA (2011) Measuring traction forces of motile dendritic cells on micropost arrays. Biophys J 101(11):2620–2628. doi:10.1016/j.bpj.2011.09.022
Soon FC, Tee SK, Youseffi M, Denyer CM (2015) Tracking traction force changes of single cells on the liquid crystal surface. Biosensors 5(1):13–24. doi:10.3390/bios5010013
Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher DE (2004) Substrate compliance versus ligand density in cell on gel responses. Biophys J 86(1):617–628. doi:10.1016/S0006-3495(04)74140-5
Reinhart-King CA, Dembo M, Hammer DA (2005) The dynamics and mechanics of endothelial cell spreading. Biophys J 89(1):676–689. doi:10.1529/biophysj.104.054320
Maloney JM, Walton EB, Bruce CM, Van Vliet KJ (2008) Influence of finite thickness and stiffness on cellular adhesion-induced deformation of compliant substrata. Phys Rev E 78(4):041923. doi:10.1103/PhysRevE.78.041923
Zhou DW, García AJ (2015) Measurement systems for cell adhesive forces. J Biomech Eng 137(2):020908. doi:10.1115/1.4029210
Ruder WC, LeDuc PR (2012) Cells gain traction in 3D. Proc Natl Acad Sci USA 109(28):11060–11061. doi:10.1073/pnas.1208617109
Tambe DT, Corey Hardin C, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EH, Zaman MH, Butler JP, Weitz DA, Fredberg JJ, Trepat X (2011) Collective cell guidance by cooperative intercellular forces. Nat Mater 10(6):469–475. doi:10.1038/nmat3025
Friedl P, Wolf K, Zegers MM (2014) Rho-directed forces in collective migration. Nat Cell Biol 16(3):208–210. doi:10.1038/ncb2923
Tambe DT, Croutelle U, Trepat X, Park CY, Kim JH, Millet E, Butler JP, Fredberg JJ (2013) Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. PLoS One 8(2):e55172. doi:10.1371/journal.pone.0055172
Murray JD, Oster GF (1984) Cell traction models for generating pattern and form in morphogenesis. J Math Biol 19(3):265–279. doi:10.1007/BF00277099
Szabó A, Mayor R (2015) Cell traction in collective cell migration and morphogenesis: The chase and run mechanism. Cell Adh Migr 9(5):380–383. doi:10.1080/19336918.2015.1019997
Varner VD, Nelson CM (2014) Cellular and physical mechanisms of branching morphogenesis. Development 141(14):2750–2759. doi:10.1242/dev.104794
Siedlik MJ, Nelson CM (2015) Regulation of tissue morphodynamics: an important role for actomyosin contractility. Curr Opin Genet Dev 32:80–85. doi:10.1016/j.gde.2015.01.002
Thompson DAW (1917) On growth and form. Cambridge University Press, Cambridge
Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE (1994) Engineering cell shape and function. Science 264(5159):696–698. doi:10.1126/science.8171320
Huang S, Ingber DE (1999) The structural and mechanical complexity of cell-growth control. Nat Cell Biol 1(5):E131–E138. doi:10.1038/13043
Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS (2005) Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci USA 102(33):11594–11599. doi:10.1073/pnas.0502575102
Chanet S, Martin AC (2014) Chapter thirteen—mechanical force sensing in tissues. In: Adam JE, Sanjay K (eds) Progress in molecular biology and translational science, vol 126. Academic Press, London, pp 317–352. doi:10.1016/B978-0-12-394624-9.00013-0
Hutson MS, Tokutake Y, Chang M-S, Bloor JW, Venakides S, Kiehart DP, Edwards GS (2003) Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science 300(5616):145–149. doi:10.1126/science.1079552
Campas O, Mammoto T, Hasso S, Sperling RA, O’Connell D, Bischof AG, Maas R, Weitz DA, Mahadevan L, Ingber DE (2014) Quantifying cell-generated mechanical forces within living embryonic tissues. Nat Meth 11(2):183–189. doi:10.1038/nmeth.2761
Rauzi M, Lenne PF (2015) Probing cell mechanics with subcellular laser dissection of actomyosin networks in the early developing Drosophila embryo. In: Nelson CM (ed) Tissue morphogenesis: methods and protocols. Springer, New York, pp 209–218
Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, Grill SW, Heisenberg C-P (2012) Forces driving epithelial spreading in zebrafish gastrulation. Science 338(6104):257–260. doi:10.1126/science.1224143
Rauzi M, Verant P, Lecuit T, Lenne P-F (2008) Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat Cell Biol 10(12):1401–1410. doi:10.1038/ncb1798
Varner VD, Nelson CM (2013) Let’s push things forward: disruptive technologies and the mechanics of tissue assembly. Integr Biol 5(9):1162–1173. doi:10.1039/C3IB40080H
Farge E (2011) Chapter eight—mechanotransduction in development. In: Michel L (ed) Current topics in developmental biology, vol 95. Academic Press, London, pp 243–265. doi:10.1016/B978-0-12-385065-2.00008-6
Mammoto T, Ingber DE (2010) Mechanical control of tissue and organ development. Development (Cambridge, England) 137(9):1407–1420. doi:10.1242/dev.024166
Rauzi M, Lenne P-F (2011) Chapter four—cortical forces in cell shape changes and tissue morphogenesis. In: Michel L (ed) Current topics in developmental biology, vol 95. Academic Press, London, pp 93–144. doi:10.1016/B978-0-12-385065-2.00004-9
Mooney JF, Hunt AJ, McIntosh JR, Liberko CA, Walba DM, Rogers CT (1996) Patterning of functional antibodies and other proteins by photolithography of silane monolayers. Proc Natl Acad Sci USA 93(22):12287–12291. doi:10.1073/pnas.93.22.12287
Alom Ruiz S, Chen CS (2007) Microcontact printing: A tool to pattern. Soft Matter 3(2):168–177. doi:10.1039/B613349E
Shen K, Qi J, Kam LC (2008) Microcontact printing of proteins for cell biology. JoVE 22:e1065. doi:10.3791/1065
Nelson CM, Inman JL, Bissell MJ (2008) Three-dimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat Protocols 3(4):674–678. doi:10.1038/nprot.2008.35
Piotrowski-Daspit AS, Tien J, Nelson CM (2016) Interstitial fluid pressure regulates collective invasion in engineered human breast tumors via Snail, vimentin, and E-cadherin. Integr Biol 8(3):319–331. doi:10.1039/C5IB00282F
Xu T, Jin J, Gregory C, Hickman JJ, Boland T (2005) Inkjet printing of viable mammalian cells. Biomaterials 26(1):93–99. doi:10.1016/j.biomaterials.2004.04.011
Murphy SV, Atala A (2014) 3D bioprinting of tissues and organs. Nat Biotech 32(8):773–785. doi:10.1038/nbt.2958
Tien J, Nelson CM, Chen CS (2002) Fabrication of aligned microstructures with a single elastomeric stamp. Proc Natl Acad Sci USA 99(4):1758–1762. doi:10.1073/pnas.042493399
Pavlovich AL, Manivannan S, Nelson CM (2010) Adipose stroma induces branching morphogenesis of engineered epithelial tubules. Tissue Eng Part A 16(12):3719–3726. doi:10.1089/ten.tea.2009.0836
Boghaert E, Gleghorn JP, Lee K, Gjorevski N, Radisky DC, Nelson CM (2012) Host epithelial geometry regulates breast cancer cell invasiveness. Proc Natl Acad Sci USA 109(48):19632–19637. doi:10.1073/pnas.1118872109
Bernard A, Renault JP, Michel B, Bosshard HR, Delamarche E (2000) Microcontact printing of proteins. Adv Mater 12(14):1067–1070. doi:10.1002/1521-4095(200007)12:14<1067:AID-ADMA1067>3.0.CO;2-M
Wilbur JL, Kumar A, Kim E, Whitesides GM (1994) Microfabrication by microcontact printing of self-assembled monolayers. Adv Mater 6(7–8):600–604. doi:10.1002/adma.19940060719
Nelson CM, Raghavan S, Tan JL, Chen CS (2003) Degradation of micropatterned surfaces by cell-dependent and -independent processes. Langmuir 19(5):1493–1499. doi:10.1021/la026178b
Lee J, Abdeen AA, Tang X, Saif TA, Kilian KA (2015) Geometric guidance of integrin mediated traction stress during stem cell differentiation. Biomaterials 69:174–183. doi:10.1016/j.biomaterials.2015.08.005
Polio SR, Smith ML (2014) Chapter 2—patterned hydrogels for simplified measurement of cell traction forces. In: Matthieu P, Manuel T (eds) Methods in cell biology, vol 121. Academic Press, London, pp 17–31. doi:10.1016/B978-0-12-800281-0.00002-6
Polio SR, Parameswaran H, Canovic EP, Gaut CM, Aksyonova D, Stamenovic D, Smith ML (2014) Topographical control of multiple cell adhesion molecules for traction force microscopy. Integr Biol 6(3):357–365. doi:10.1039/C3IB40127H
Vedula SRK, Leong MC, Lai TL, Hersen P, Kabla AJ, Lim CT, Ladoux B (2012) Emerging modes of collective cell migration induced by geometrical constraints. Proc Natl Acad Sci USA 109(32):12974–12979. doi:10.1073/pnas.1119313109
Yang MT, Fu J, Wang Y-K, Desai RA, Chen CS (2011) Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nat Protocols 6(2):187–213. doi:10.1038/nprot.2010.189
Han SJ, Bielawski KS, Ting LH, Rodriguez ML, Sniadecki NJ (2012) Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys J 103(4):640–648. doi:10.1016/j.bpj.2012.07.023
Wang N, Ostuni E, Whitesides GM, Ingber DE (2002) Micropatterning tractional forces in living cells. Cell Motil Cytoskeleton 52(2):97–106. doi:10.1002/cm.10037
Gomez EW, Chen QK, Gjorevski N, Nelson CM (2010) Tissue geometry patterns epithelial–mesenchymal transition via intercellular mechanotransduction. J Cell Biochem 110(1):44–51. doi:10.1002/jcb.22545
Rolli CG, Nakayama H, Yamaguchi K, Spatz JP, Kemkemer R, Nakanishi J (2012) Switchable adhesive substrates: revealing geometry dependence in collective cell behavior. Biomaterials 33(8):2409–2418. doi:10.1016/j.biomaterials.2011.12.012
Raghavan S, Desai RA, Kwon Y, Mrksich M, Chen CS (2010) Micropatterned dynamically adhesive substrates for cell migration. Langmuir 26(22):17733–17738. doi:10.1021/la102955m
Jiang X, Ferrigno R, Mrksich M, Whitesides GM (2003) Electrochemical desorption of self-assembled monolayers noninvasively releases patterned cells from geometrical confinements. J Am Chem Soc 125(9):2366–2367. doi:10.1021/ja029485c
Gomez EW, Nelson CM (2011) Lithographically defined two- and three-dimensional tissue microarrays. In: Khademhosseini A, Suh K-Y, Zourob M (eds) Biological microarrays: methods and protocols. Humana Press, Totowa, pp 107–116. doi:10.1007/978-1-59745-551-0_5
Manivannan S, Gleghorn JP, Nelson CM (2012) Engineered tissues to quantify collective cell migration during morphogenesis. In: Michos O (ed) Kidney development: methods and protocols. Humana Press, Totowa, pp 173–182. doi:10.1007/978-1-61779-851-1_16
Zhu W, Nelson CM (2013) PI3K regulates branch initiation and extension of cultured mammary epithelia via Akt and Rac1 respectively. Dev Biol 379(2):235–245. doi:10.1016/j.ydbio.2013.04.029
Lee K, Gjorevski N, Boghaert E, Radisky DC, Nelson CM (2011) Snail1, Snail2, and E47 promote mammary epithelial branching morphogenesis. EMBO J 30(13):2662–2674. doi:10.1038/emboj.2011.159
Pavlovich AL, Boghaert E, Nelson CM (2011) Mammary branch initiation and extension are inhibited by separate pathways downstream of TGFβ in culture. Exp Cell Res 317(13):1872–1884. doi:10.1016/j.yexcr.2011.03.017
Nelson CM, VanDuijn MM, Inman JL, Fletcher DA, Bissell MJ (2006) Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science 314(5797):298–300. doi:10.1126/science.1131000
Gjorevski N, Nelson CM (2010) Endogenous patterns of mechanical stress are required for branching morphogenesis. Integr Biol 2(9):424–434. doi:10.1039/C0IB00040J
Legant WR, Chen CS, Vogel V (2012) Force-induced fibronectin assembly and matrix remodeling in a 3D microtissue model of tissue morphogenesis. Integr Biol 4(10):1164–1174. doi:10.1039/C2IB20059G
Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS (2009) Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proc Natl Acad Sci USA 106(25):10097–10102. doi:10.1073/pnas.0900174106
Sakar MS, Eyckmans J, Pieters R, Eberli D, Nelson BJ, Chen CS (2016) Cellular forces and matrix assembly coordinate fibrous tissue repair. Nat Commun 7:11036. doi:10.1038/ncomms11036
Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM (2009) Self-organization of engineered epithelial tubules by differential cellular motility. Proc Natl Acad Sci USA 106(35):14890–14895. doi:10.1073/pnas.0901269106
Théry M (2010) Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci 123(24):4201. doi:10.1242/jcs.075150
Jones CAR, Cibula M, Feng J, Krnacik EA, McIntyre DH, Levine H, Sun B (2015) Micromechanics of cellularized biopolymer networks. Proc Natl Acad Sci USA 112(37):E5117–E5122. doi:10.1073/pnas.1509663112
Kim J, Jones CAR, Groves NS, Sun B (2016) Three-dimensional reflectance traction microscopy. PLoS One 11(6):e0156797. doi:10.1371/journal.pone.0156797
Notbohm J, Lesman A, Tirrell DA, Ravichandran G (2015) Quantifying cell-induced matrix deformation in three dimensions based on imaging matrix fibers. Integr Biol 7(10):1186–1195. doi:10.1039/C5IB00013K
Cost AL, Ringer P, Chrostek-Grashoff A, Grashoff C (2015) How to measure molecular forces in cells: a guide to evaluating genetically-encoded FRET-based tension sensors. Cell Mol Bioeng 8(1):96–105. doi:10.1007/s12195-014-0368-1
Gayrard C, Borghi N (2016) FRET-based molecular tension microscopy. Methods 94:33–42. doi:10.1016/j.ymeth.2015.07.010
de Souza N (2012) Pulling on single molecules. Nat Meth 9(9):873–877. doi:10.1038/nmeth.2149
Liu B, Kim T-J, Wang Y (2010) Live cell imaging of mechanotransduction. J R Soc. Interface 7(Suppl 3):S365–S375. doi:10.1098/rsif.2010.0042.focus
Leidel C, Longoria RA, Gutierrez FM, Shubeita GT (2012) Measuring molecular motor forces in vivo: implications for tug-of-war models of bidirectional transport. Biophys J 103(3):492–500. doi:10.1016/j.bpj.2012.06.038
Zhang Y, Ge C, Zhu C, Salaita K (2014) DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat Commun. doi:10.1038/ncomms6167
Morimatsu M, Mekhdjian AH, Chang AC, Tan SJ, Dunn AR (2015) Visualizing the interior architecture of focal adhesions with high-resolution traction maps. Nano Lett 15(4):2220–2228. doi:10.1021/nl5047335
Laakso JM, Lewis JH, Shuman H, Ostap EM (2008) Myosin I can act as a molecular force sensor. Science 321(5885):133–136. doi:10.1126/science.1159419
Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, Chen CS (2014) A DNA-based molecular probe for optically reporting cellular traction forces. Nat Meth 11(12):1229–1232. doi:10.1038/nmeth.3145
Stabley DR, Jurchenko C, Marshall SS, Salaita KS (2012) Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat Meth 9(1):64–67. doi:10.1038/nmeth.1747
Zeinab A-R, Andrew EP (2013) Cross talk between matrix elasticity and mechanical force regulates myoblast traction dynamics. Phys Biol 10(6):066003. doi:10.1088/1478-3975/10/6/066003
Haase K, Pelling AE (2015) Investigating cell mechanics with atomic force microscopy. J R Soc Interface 12(104):20140970. doi:10.1098/rsif.2014.0970
Sniadecki NJ, Anguelouch A, Yang MT, Lamb CM, Liu Z, Kirschner SB, Liu Y, Reich DH, Chen CS (2007) Magnetic microposts as an approach to apply forces to living cells. Proc Natl Acad Sci USA 104(37):14553–14558. doi:10.1073/pnas.0611613104
Mejean CO, Schaefer AW, Millman EA, Forscher P, Dufresne ER (2009) Multiplexed force measurements on live cells with holographic optical tweezers. Opt Express 17(8):6209–6217. doi:10.1364/OE.17.006209
van der Horst A, Forde NR (2008) Calibration of dynamic holographic optical tweezers for force measurements on biomaterials. Opt Express 16(25):20987–21003. doi:10.1364/OE.16.020987
Ladoux B (2009) Biophysics: Cells guided on their journey. Nat Phys 5(6):377–378. doi:10.1038/nphys1281
Franck C, Maskarinec SA, Tirrell DA, Ravichandran G (2011) Three-dimensional traction force microscopy: a new tool for quantifying cell–matrix interactions. PLoS One 6(3):e17833. doi:10.1371/journal.pone.0017833
Rape AD, Guo W-H, Wang Y-L (2011) The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials 32(8):2043–2051. doi:10.1016/j.biomaterials.2010.11.044
Polio SR, Rothenberg KE, Stamenović D, Smith ML (2012) A micropatterning and image processing approach to simplify measurement of cellular traction forces. Acta Biomater 8(1):82–88. doi:10.1016/j.actbio.2011.08.013
Cvetkovic C, Raman R, Chan V, Williams BJ, Tolish M, Bajaj P, Sakar MS, Asada HH, Saif MTA, Bashir R (2014) Three-dimensionally printed biological machines powered by skeletal muscle. Proc Natl Acad Sci USA 111(28):10125–10130. doi:10.1073/pnas.1401577111
Sung JH, Yu J, Luo D, Shuler ML, March JC (2011) Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 11(3):389–392. doi:10.1039/C0LC00273A
Acknowledgements
Work from the authors’ group was supported in part by grants from the NIH (GM083997, HL110335, HL118532, HL120142, and CA 187692), the NSF (CMMI-1435853), the David and Lucile Packard Foundation, the Alfred P. Sloan Foundation, the Camille and Henry Dreyfus Foundation, and the Burroughs Wellcome Fund. M.J.S. was supported in part by the NSF Graduate Research Fellowship Program. C.M.N. was supported in part by a Faculty Scholars Award from the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nerger, B.A., Siedlik, M.J. & Nelson, C.M. Microfabricated tissues for investigating traction forces involved in cell migration and tissue morphogenesis. Cell. Mol. Life Sci. 74, 1819–1834 (2017). https://doi.org/10.1007/s00018-016-2439-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2439-z