Abstract
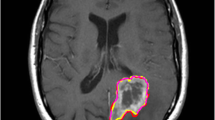
The automated segmentation of brain tumours utilizing multimodal magnetic resonance imaging (MRI) is crucial in researching and monitoring disease progression. To aid in distinguishing gliomas into intertumoural classes, efficient and precise segmentation methods are utilized to differentiate gliomas into intratumourally categorized types. Deep learning algorithms outperform classical context-based computer vision techniques in circumstances that need the segmentation of objects into categories. Convolutional neural networks (CNNs) are extensively used in medical image segmentation, and they have significantly improved the accuracy of brain tumour segmentation in the present generation. Specifically, this research introduces a residual network (ResNet), a blend of two segmentation networks that employ a primary but simple combinative method to provide better and more accurate predictions. After each model was trained on the BraTS-20 challenge data, it was analysed to yield segmentation results. Among the different methodologies examined, ResNet produced the most accurate results compared to U-Net and was thus chosen and organized in many ways to arrive at the final forecast on the validation set. The ensemble acquired dice scores of 0.80 and 0.85 for the augmentation of the tumour, total cancer, and tumour core, respectively, demonstrating more excellent performance than the present technology in use.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sauli R, Akil M, Kachori R, et al (2018) Fully automatic brain tumour segmentation using end-to-end incremental deep neural networks in MRI images. Comput Methods Programs Biomed 166:39–49
Goetz M et al (2015) DALSA: domain adaptation for supervised learning from sparsely annotated MR images. IEEE Trans Med Imaging 35(1):184–196
Farahani K, Menze B, Reyes M (2014) Brats 2014 challenge manuscripts. http//www. Brain tumour segmentation. Org
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828
Hinton GE, Osindero S, Teh Y-W (2006) A fast learning algorithm for deep belief nets. Neural Comput 18(7):1527–1554
Bengio Y, Lamblin P, Popovici D, Larochelle H (2007) Greedy layer-wise training of deep networks. Adv Neural Inf Proc Syst 153–160
Lee H, Ekanadham C, Ng AY (2008) Sparse deep belief net model for visual area V2. Adv Neural Inf Proc Syst 873–880
Wang G, Li W, Vercauteren T, Ourselin S (2019) Automatic brain tumour segmentation based on cascaded convolutional neural networks with uncertainty estimation. Front Comput Neurosci 13:56
Mukherjee P, Mukherjee A (2019) Advanced processing techniques and secure architecture for sensor networks in ubiquitous healthcare systems. Sens Health Monit 3–29 (Elsevier)
Bauer S, Wiest R, Nolte LP, Reyes M (2013) A survey of MRI-based medical image analysis for brain tumour studies
Leece R, Xu J, Ostrom QT, Chen Y, Kruchko C, Barnholtz-Sloan JS (2017) Global incidence of malignant brain and other central nervous system tumours by histology, 2003–2007. Neuro Oncol 19(11):1553–1564
Dolecek TA, Propp JM, Stroup NE, Kruchko C (2012) CBTRUS statistical report: primary brain and central nervous system tumours diagnosed in the United States in 2005—2009. Neuro Oncol 14(5):1–49
Louis DN et al (2016) The 2016 World Health Organization classification of tumours of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Hlama, S.T., Ghanim, S.A., Dakheel, H.R., Mohammed, S.H. (2023). Deep Neural Ideal Networks for Brain Tumour Image Segmentation. In: Khanna, A., Polkowski, Z., Castillo, O. (eds) Proceedings of Data Analytics and Management . Lecture Notes in Networks and Systems, vol 572. Springer, Singapore. https://doi.org/10.1007/978-981-19-7615-5_27
Download citation
DOI: https://doi.org/10.1007/978-981-19-7615-5_27
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-7614-8
Online ISBN: 978-981-19-7615-5
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)