Abstract
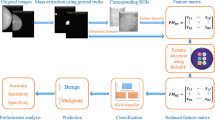
Feature extraction and selection are very important stages in pattern recognition and computer vision solutions with far-reaching effects on their performance. In computer-aided diagnosis (CADx) systems, efficiency is affected by its subjectivity to the accuracy of the region of interest (ROI) extraction technique, which is largely dependent on the features extracted. Optimization algorithms are often used to improve the selection of discriminative features which thereby leads to improve accuracy of the CADx systems. This work considers the effects of optimizing selected features in the performance of breast tissue characterization in mammograms. It uses Whale Optimization Algorithm (WOA) to optimize Otsu fitness function of Gray Level Co-occurrence Matrix (GLCM) in extracting the region of interest (ROI). The extracted features were classified into BIRADS scales 1, 2 and 5 using Multiclass Support Vector Machine (MSVM). The performance of the developed algorithm was evaluated using specificity, sensitivity as well as accuracy and compared with other techniques namely Texture Signature (TS), Pixel-Based Morphological (PBM), Natural Language Processing (NLP), and Interactive Data Language (IDL). The result of the developed WOA-Otsu-GLCM-MSVM CADx algorithm for specificity, sensitivity, and accuracy are 96%, 92% and 94%, respectively. The developed algorithm gave an accuracy of 94.4% as against 81.0%, 85.7%, 93.0% and 82.5% for TS, PBM, NLP and IDL methods, respectively. The characterization of the breast tumour using the developed CADx algorithm performed better compared with the conventional methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
El Aziz, M. A., Ewees, A. A., Hassanien, A. E., Mudhsh, M., & Xiong, S. (2018). Multi-objective whale optimization algorithm for multilevel thresholding segmentation. In Advances in soft computing and machine learning in image processing (Studies in computational intelligence) (Vol. 730, pp. 23–39). Springer.
Hassanpour, H., Samadiani, N., & Salehi, S. M. (2015). Using morphological transforms to enhance the contrast of medical images. The Egyptian Journal of Radiology and Nuclear Medicine, 46(2), 481–489.
Wang, C., Brentnall, A. R., Cuzick, J., Harkness, E. F., Evans, D. G., & Astley, S. (2017). A novel and fully automated mammographic texture analysis for risk prediction: Results from two case-control studies. Breast Cancer Research, 19, 114.
Priyanka, D., & Chinmay, C. (2021). Application of AI on post pandemic situation and lesson learn for future prospects. Journal of Experimental & Theoretical Artificial Intelligence, 1, 1–24.
Chinmay, C. (2019). Performance analysis of compression techniques for chronic wound image transmission under smartphone-enabled tele-wound network. International Journal of E-Health and Medical Communications (IJEHMC), 10(2), 1–15.
Saidin, N., Ngah, K. U., Shuaib, L. I., & Sakim, H. A. M. Segmentation of breast regions in mammogram based on density: A review. Imaging & Computational Intelligence Group (ICI).
Chakravarthi, R., Nandhitha, N. M., Roslin, S. E., & Selvarasu, N. (2016). Tumour extraction from breast mammographs through hough transform and DNN hybrid segmentation technique. Biomedical Research, 27(4), 1188–1193.
Krishnan, M., Chinmay, C., Banerjee, S., Chakraborty, C., & Ray, A. K. (2009). Statistical analysis of mammographic features and its classification using support vector machine. Expert Systems with Applications, 37, 470–478.
Kaur, P., Singh, G., & Kaur, P. (2019). Intellectual detection and validation of automated mammogram breast cancer images by multi-class SVM using deep learning classification. Informatics in Medicine Unlocked, 16, 100151.
Ganesan, K., Acharya, U. R., Chua, C. K., Min, L. C., Abraham, K. T., & Kwan-Hoong, N. (2013). Computer-aided breast cancer detection using mammograms: A review. IEEE Reviews in Biomedical Engineering, 6, 77–98.
Pavol, Z., Peter, K., Karol, K., Zuzana, D., Hubert, P., Tibor, B., Erik, K., Marek, S., Alena, L., Dominika, V., Tatiana, K., Igor, S., Veronica, H., Jan, B., Zuzana, L., Dietrich, B., Mariusz, A., Walther, K., Jan, D., & Olga, G. (2019). Why the gold standard approach by mammography demands extension by multiomics? Application of liquid Biopsis miRNA profiles to breast cancer disease management. International Journal of MolecularSciences, 20(12), 2878.
Siegel, R. L., Miller, K. D., Fedewa, S. A., Ahnen, D. J., Meester, R. G. S., Barz, A., & Jemal, A. (2017). Colorectal cancer statistics. CA: a Cancer Journal for Clinicians, 67(3), 177–193.
Ford, D., Easton, D. F., Stratton, M., Narod, S., Goldgar, D., Devilee, P., Bishop, D. T., Weber, B., Lenoir, G., Chang-Claude, J., Sobol, H., Teare, M. D., Struewing, J., Arason, A., Scherneck, S., Peto, J., Rebbeck, T. R., Tonin, P., Neuhausen, S., … Eyfjord, J. (1998). Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. American Journal of Human Genetics, 62, 676–689.
Kallenberg, M., Petersen, K., Nielsen, M., Ng, A. Y., Diao, P., Igel, C., Vachon, C. M., Holland, K., Winkel, R. R., Karssemeijer, N., & Lillholm, M. (2016). Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Transactions on Medical Imaging, 35(5), 1322–1331.
Sayed, G. I., Darwish, A., Hassanien, A. E., & Pan, J.-S. (2017). Breast cancer diagnosis approach based on meta-heuristic optimization algorithm inspired by the bubble-net hunting strategy of whales. In Genetic and evolutionary computing (Advances in intelligent systems and computing). Springer.
Nasiri, J., & Khiyaban, F. M. (2018). A whale optimization algorithm (WOA) approach for clustering. Cogent Mathematics & Statistic, 5, 1483565.
Yogapriya, J., Saravanabhavan, C., & Ila, V. (2018). Medical image retrieval system using local binary patterns,whale optimization & relevance vector machine algorithms. Tagajuornal, 14, 3164–3191.
Adepoju, T. M., Ojo, J. A., Omidiora, E. O., & Olabiyisi, O. S. (2015). Pixel-based morphological technique for breast tumour detection. International Journal of Scientific & Engineering Research, 6(6), 1416–1420.
ACS. (2017). Breast cancer early detection and diagnosis.
Jalalian, A., Mashohor, S., Mahmud, R., Karasfi, B., Saripan, M. I. B., & Ramli, A. R. B. (2017). Review article: Foundation and methodologies in computer-aided diagnosis systems for breast cancer detection. EXCLI Journal, 16, 113–137.
Saslow, D., Boetes, C., & Burke, W. (2007). American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA: A Cancer Journal for Clinicians, 57(2), 75–89.
Raj, J. R., Rahman, S. M. K., & Anand, S. (2016). Preliminary evaluation of differentiation of benign and malignant breast tumors using non-invasive diagnostic modalities. Biomedical Research, 27(3), 596–603.
Adeyemo, T. T., Adepujo, T. M., Sobowale, A. A., Oyediran, M. O., Omidiora, E. O., & Olabiyisi, S. O. (2017). Feature extraction techniques for mass detection in digital mammogram (review). Journal of Scientific Research & Reports, 17(1), 1–11.
Shallu, R. M. (2018). Breast cancer histology images classification: Training from scratch or transfer learning? ICT Express, 4, 247–254.
Rakhlin, A., Shvets, A., Iglovikov, V., & Kalinin, A. A. (2018). Deep convolutional neural networks for breast cancer histology image analysis. In Lecture notes in computer science (Vol. 10882, pp. 737–744). Springer.
Arau’jo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P., Eloy, C., et al. (2017). Classification of breast cancer histology images using convolutional neural networks. PLoS One, 12(6), e0177544.
Tahir, M., Muhammad, A. M. O., Min, B. L., & Kang, R. P. (2020). Artificial intelligence-based mitosis detection in breast cancer histopathology images using faster R-CNN and Deep CNNs. Journal of Clinical Medicine, 9, 749.
Alkassar, S., Jebur, B. A., Abdullah, M. A. M., Al-Khalidy, J. H., & Chambers, J. A. Going deeper: Magnification invariant approach for breast cancer classification using histopathological images. IET Computer Vision, 15, 151–164.
Adepoju, T. M., Adeyemo, T. T., Fagbola, T. M., Omidiora, E. O., & Olabiyisi, S. O. (2016). Histogram normalization technique for preprocessing of digital mammographic images. LAUTECH Journal of Engineering and Technology, 10(1), 82–87.
Corinne, B., Salma, A., Kim, V. N., Daniel, V., Clarisse, D., & Robert, S. (2007). BIRADSTM classification in mammography. European Journey of Radiology, 61, 192–194.
Rampun, P., Morrow, J., Scotney, B. W., & Wang, H. (2020). Breast density classification in mammograms: An investigation of encoding techniques in binary-based local patterns. Computers in Biology and Medicine, 122, 1–18.
Aghdam, H., Puig, D., & Solanas, A. (2013). A probabilistic approach for breast boundary extraction in mammograms. Computational and Mathematical Methods in Medicine, 10, 17–26.
Suguna, S. K., & Ranganathan, R. (2017). A new Evolutionanary – Based optimization algorithm for mammogram image processing. International Journal of Pure and Applied Mathematics, 117(17), 241–247.
Cruz, C. F. (2011). Automatic analysis of mammography images: Enhancement and segmentation techniques. Port University.
Karakoyun, M., Baykan, N. A., & Hacibeyoglu, M. (2017). Multi-level thresholding for image segmentation with swarm optimization algorithms. International Research Journal of Electronics & Computer Engineering, 3(3), 654–658.
Kumar, T. G., Murugan, D., & Manish, T. I. (2018). An analysis on road extraction from satellite image using Otsu method and genetic algorithm techniques. WSEAS Transactions on Computers, 17, 42–51.
Mirjalili, S., & Lewis, A. (2016). The whale optimization algorithm. Advances in Engineering Software, 95, 51–67.
Zidan, M., Hassanien, A. E., Hefny, H. A., & Houseni, M. (2017). Liver segmentation in MRI images based on whale optimization algorithm. Multimedia Tools and Applications, 76, 24931–24954.
Ozturk, Ş., Akdemir, B., Ozkaya, U., & Seyfi, L. (2017). Soft tissue sacromas segmentation using optimized otsu thresholding algorithms. International Journal of Engineering Technology, Management and Applied Sciences, 5, 9.
Mafarja, M. M., & Mirjalili, S. (2017). Hybrid Whale Optimization Algorithm with simulated annealing for feature selection. Neurocomputing, 260, 302–312.
Kowsalya, S., & Priyaa, D. S. (2015). A survey on diagnosis methods of breast cancer using mammography. International Journal of Engineering Technology Science and Research, 2, 100–107.
Guyon, I., & Elisseeff, A. (2003). Special issue on variable and feature selection. Journal of Machine Learning Research, 3, 1157–1182.
Mohanaiah, P., Sathyanarayana, P., & GuruKumar, L. (2013). Image texture feature extraction using GLCM approach. International Journal of Scientific and Research Publications, 3(5), 1–5.
Punithavathi, V., & Devakumari, D. (2020) Detection of breast lesion using improved GLCM feature based extraction in mammogram images.
Jagadesh, B. N., & Kumari, L. K. (2021). A GLCM based feature extraction in mammogram images using machine learning algorithms. International Journal of Current Research and Review, 13(5), 145–149.
Mahdi, F. (2015). Application of GLCM technique on mammograms for early detection of breast cancer. Journal of Babylon University/Pure and Applied Sciences, 23(2), 885–890.
Chinmay, C. (2019). Computational approach for chronic wound tissue characterization. Elsevier: Informatics in Medicine Unlocked, 17, 1–10.
Mryka, H. B. (2017). GLCM texture: A tutorial. University of Calgar.
Ojo, J. E., Adepoju, T. M., Omidiora, E. O., Olabiyisi, O. S., & Bello, O. T. (2014). Pre-processing method for extraction of pectoral muscle and removal of artefacts in mammogram. IOSR Journal of Computer Engineering (IOSR-JCE), 16(3), 06–09.
Adepoju, T. M., Ojo, J. A., Omidiora, E. O., Olabiyisi, O. S., & Bello, O. T. (2015). Detection of tumour based on breast tissue categorization. British Journal of Applied Science & Technology, 11(5), 1–12.
Mohamed, S. E., Wahbi, T. M., & Sayed, M. H. (2018). Automated detection and classification of breast cancer using mammography images. International Journal of Science, Engineering and Technology Research (IJSETR), 7, 4.
S. M. Castro, E. Tseytlin, O. Medvedeva, . K. Mitchell, S. Visweswaran, T. Bekhuis and R. S. Jacobson, "Automated annotation and classification of BI-RADS assessment from radiology reports, Journal of Biomedical Informatics" 67, 177–187, 2017.
He, W., Denton, E. R., & Zwiggelaar, R. (2010). Mammographic image segmentation and risk classification using a novel texture signature based methodology. IWDM, 6136, 526–533.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ojo, J.A., Bello, T.O., Idowu, P.O., Solomon, I.D. (2022). Optimal Feature Selection for Computer-Aided Characterization of Tissues: Case Study of Mammograms. In: Chakraborty, C., Khosravi, M.R. (eds) Intelligent Healthcare. Springer, Singapore. https://doi.org/10.1007/978-981-16-8150-9_3
Download citation
DOI: https://doi.org/10.1007/978-981-16-8150-9_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-8149-3
Online ISBN: 978-981-16-8150-9
eBook Packages: Computer ScienceComputer Science (R0)