Abstract
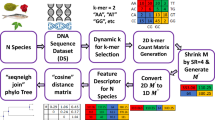
Bioinformatics, which is now a well-known field of study, originated in the context of biological sequence analysis. Now in bioinformatics, we found a wide range of applications in the domain of molecular biology, which focuses on the analysis of molecules. The most important thing in the domain of DNA sequence analysis is sequence analysis. Sequence analysis algorithms are designed to identify the origin of viruses, the amount of similarity between viruses/species, and also the amount of mutation occurs in the viruses/species. In this paper, we adopt a new approach in DNA sequence similarity analysis that differs from the attempts made in the past. Our method is highly effective, accurate, and reliable to identify the similarity with point mutation. Our goal is to reduce the time complexity of similarity analysis. We find the amount of similarity among different species. It takes \(\mathcal {O}(n)\) time to identify the amount of similarity with a point mutation, which is faster than the other alignment algorithms including the Needleman-Wunsch (NW) algorithm, FOGSAA, MEGA, and BLASTN.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Needleman, S.B., Wunsch, C.D.: A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443–53 (1970)
Smith, T.F., Waterman, M.S.: Identification of common molecular subsequences. J. Mol. Biol. 147(1), 195–197 (1981)
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J.: Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
Menlove, K.J., Clement, M., Crandall, K.A.: Similarity searching using BLAST. J. Bioinform. DNA Sequence Anal. Methods Mol. Biol. 537 (2009). ISBN 978-1-59745-251-9
Pearson, W.R., Lipman, D.J.: Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 85, 2444–2448 (1988)
Huang, X., Chao, K.: A generalized global alignment algorithm. Bioinformatics 19, 228–233 (2003)
Bray, N., Dubchak, I., Pachter, L.: AVID: a global alignment program. Genome Res. 13, 97–102 (2003)
Rizk, G., Lavenier, D.: GASSST: global alignment short sequence search tool. Bioinformatics 26, 2534–2540 (2010)
Yu, J.-F., Wang, J.-H., Sun, X.: Analysis of similarities/dissimilarities of DNA sequence based on a novel graphical Representation. MATCH Commun. Math. Comput. Chem. (2010)
Qi, X., Qin, W., Zhang, Y., Fuller, E., Zhang, C.-Q.: A novel model for DNA sequence similarity analysis based on graph theory evolutionary bioinformatics 7, 149–158 (2011)
Zhang, Y.: A simple method to construct the similarity matrices of DNA sequence. Match Commun. Comput. Chem. 60, 313–24 (2008)
Zhang, Z., Wang, S., Zhang, X., Zhang, Z.: Similarity analysis of DNA sequences based on a compact representation. IEEE (2010). 978-1-4244-6439
Roy, A., Raychaudhury, C., Nandy, A.: Novel techniques of graphical representation and analysis of DNA sequences—A review, vol. 23, p. 17 (1998). https://doi.org/10.1007/bf02728525
Hamming, R.W.: Error detecting and error correcting codes. Bell Syst. Tech. J. 29(2), 147–160 (1950). ISSN 0005-8580. https://doi.org/10.1002/j.1538-7305.1950.tb00463.x
Chakraborty, A., Bandyopadhyay, S.: FOGSAA: fast optimal global sequence alignment algorithm. Sci. Rep. 3, 1746 (2013). https://doi.org/10.1038/srep01746
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Mondal, P., Basuli, K. (2022). OAIPM: Optimal Algorithm to Identify Point Mutation Between DNA Sequences. In: Mandal, J.K., Buyya, R., De, D. (eds) Proceedings of International Conference on Advanced Computing Applications. Advances in Intelligent Systems and Computing, vol 1406. Springer, Singapore. https://doi.org/10.1007/978-981-16-5207-3_33
Download citation
DOI: https://doi.org/10.1007/978-981-16-5207-3_33
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-5206-6
Online ISBN: 978-981-16-5207-3
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)