Abstract
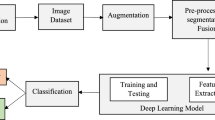
Recently, computer-aided diagnosis methods based on machine learning, mainly using Deep Leaning, have been studied and developed very rapidly. Especially image recognition based on Convolutional Neural Networks showed high accuracy in diagnosis problems when they are given the huge amount of training data. Those methods are not only helpful for classification but also useful for feature extraction from given images. Here we introduce a new classification method to find the features of tumor tissues from histopathology images by unsupervised clustering based on Information Maximization Self-Augmented Training. Moreover, to evaluate fibrosis and classify tumor cells, we used histopathological images with different staining methods as concatenated inputs. Using this approach, we can quantify integrated features based on multimodal imaging using deep learning. In this study, we analyzed pathological images of pancreas cancers and optimized to classify the patches of the images into the categorize with different features, which are consistent with annotation of the medical doctors. It can also provide a map to visualize the probability where cell types are categorized into specific classes according to the given pathological images.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Van Ginneken B, Romeny BT, Viergever MA. Computer-aided diagnosis in chest radiography: a survey. IEEE Trans Med Imaging. 2001 Dec;20(12):1228–41.
Doi K, MacMahon H, Katsuragawa S, Nishikawa RM, Jiang Y. Computer-aided diagnosis in radiology: potential and pitfalls. Eur J Radiol. 1999;31(2):97–109.
Farjam R. Soltanian, Zadeh H, Jafari, Khouzani K, Zoroofi RA. An image analysis approach for automatic malignancy determination of prostate pathological images. Cytometry Part B: Clinical Cytometry: The Journal of the International Society for Analytical Cytology. 2007 Jul;72(4):227–40.
Hu W, Miyato T, Tokui S, Matsumoto E, Sugiyama M. Learning discrete representations via information maximizing self-augmented training. arXiv preprint arXiv:1702.08720. 2017 Feb 28.CNN.
Lee JW, Komar CA, Bengsch F, Graham K, Beatty GL. Genetically engineered mouse models of pancreatic Cancer: the KPC model (LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre), its variants, and their application in Immunooncology drug discovery. Current protocols in pharmacology. 2016 Jun;73(1):14–39.
Asano K, Ono N, Iwamoto C, Ohuchida K, Shindo K, and Kanaya S, “Feature extraction and Cluster analysis of Pancreatic Pathological Image Based on Unsupervised Convolutional Neural Network,” 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 2018, pp. 2738–2740, https://doi.org/10.1109/BIBM.2018.8621323.
Hontani H, et al. “Registration between histopathological images with different stains and an MRI image of pancreatic cancer tumor.” International Forum on Medical Imaging in Asia 2019. Vol. 11050. International Society for Optics and Photonics, 2019.
Mikołajczyk A, Grochowski M. Data augmentation for improving deep learning in image classification problem. In 2018 international interdisciplinary PhD workshop (IIPhDW) 2018 May 9 (pp. 117–122). IEEE.
Miyato T, Maeda SI, Koyama M, Ishii S. Virtual adversarial training: a regularization method for supervised and semi-supervised learning. IEEE Trans Pattern Anal Mach Intell. 2018 Jul 23;41(8):1979–93.
Bridle JS, Heading AJR, MacKay DJC. Unsupervised classifiers, mutual information and 'Phantom targets. Adv Neural Inf Proces Syst. 1992;4:1096–101.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Ono, N., Iwamoto, C., Ohuchida, K. (2022). Construction of Classifier of Tumor Cell Types of Pancreas Cancer Based on Pathological Images Using Deep Learning. In: Hashizume, M. (eds) Multidisciplinary Computational Anatomy. Springer, Singapore. https://doi.org/10.1007/978-981-16-4325-5_17
Download citation
DOI: https://doi.org/10.1007/978-981-16-4325-5_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-4324-8
Online ISBN: 978-981-16-4325-5
eBook Packages: MedicineMedicine (R0)