Abstract
One of the most serious viral outbreaks of the decade, infecting humans, originated from the city of Wuhan, China, by the end of December 2019, has left the world shaken up. It is the successor infection of severe acute respiratory syndrome coronavirus (SARS-CoV) named as SARS-CoV-2 causing a disease called as COVID-19 (Coronavirus disease-19). Being one of the most severe diseases in terms of transmission, this disease agitates the immune system of an individual quite disturbingly which at times leads to death, which is why it has become the need of the hour to step forward to extensively involve in understanding the genetics, pathogenesis, and immunopathology of SARS-CoV-2 in order to design drugs to treat or to design a vaccine to prevent. In this chapter, we have tried to review and summarize the studies done so far to understand the host–pathogen relationship and the host immune response during COVID-19 infection. One of the recent developments regarding the understanding of SARS-CoV-2 infection is the mechanism of immune evasion involved during the pathogenesis and cytokine storm syndrome during infection in the patient against which a drug called as Hydroxychloroquine has been designed. Comprehensively, we have tried to give an immunological insight into the SARS-CoV-2 infection in order to understand the possible outcome for any therapeutic advancement.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
With such an avalanche of reported cases of CoVID-19 (Coronavirus disease-19) from all across the globe, the family of Coronavirus has pretty nearly brought the entire human race under its feet since late December 2019 (Khan et al. 2020). “Corona” loosely meaning “halo” or “crown” in Latin refers to the structure seen by the capsid and RNA. “Coronavirus” was actually termed during the imaging of the viral family Coronaviridae, due to the circular shape of the virus itself (Li 2016). CoVID-19 is caused by the novel strain of Coronavirus named as SARS-CoV-2 due to its homology with SARS infection and has been declared as a pandemic by the WHO (World Health Organization) on March 2020 (World Health Organization 2020). This novel strain of CoV debuted into the human host causing severe pneumonia and respiratory disorders (Wang et al. 2020). SARS-CoV-2 has managed to rattle the host immune system very successfully. Reports suggest that, analogous to its predecessor (SARS-CoV), who belongs to the same genera and family, the origin of SARS-CoV-2 has also been reported to be from the species of bats (Li et al. 2020). This strain of coronavirus is highly contagious, which is the root cause of CoVID-19 being spread rapidly and causing higher number of casualties ever since the first case has been reported.
5.2 Family of Coronaviruses
Coronaviruses (CoVs) are a group of viruses that belong to the family Coronaviridae, infecting humans along with other species (not every CoVs), and are respiratory illness causing viruses. The first encounter with the CoV was seen in 1960s, which was named as HCoV-OC43 and HCoVs 229E (Drosten et al. 2003). Until late December 2019, six such groups of CoVs were being known: HCoV-OC43, HCoV-HKU1, HCoV-229E, SARS-CoV, HCoV-NL63, and MERS-CoV are among them. CoVs are categorized under four genera: Alpha CoVs (HCoV-NL63, HCoV-229), Beta CoVs (HCoV-OC43, SARS-CoV, HCoV-HKU1, and MERS-CoV), Gamma CoVs, and Delta CoVs (Fehr and Perlman 2015).
5.3 Entry of CoV into Host Cells
Coronaviruses uses a very spiky-shaped protein, S protein, to infect a cell by binding to the membrane of the cell. COVID-19 (SARS-CoV-2) and SARS-CoV share a receptor-binding unit whose domain structure is similar, suggesting that COVID-19 (SARS-CoV-2) uses ACE2 receptor in humans for infection (Yan et al. 2020). The spike protein binds to this ACE2 receptor on the host cell surface and gets pinched inside the host cell. Studies has been conducted which shows the role of an enzyme Furin present in the host cells, plays a crucial role in SARS-CoV-2 entry, and can be a distinguishing feature defining the severity of SARS-CoV-2, since it is absent in SARS-CoV (Walls et al. 2020). This enzyme activates SARS-CoV-2 whereas the SARS-CoV and MERS-CoV during entrance into the host cell do not encounter this activated site. Since furin is expressed in various human organs such as the lungs, small intestine, and liver, the infection in human has been seen to be very vigorous and can be predicted to be potentially infecting multiple human organs. This site could possibly affect the transmission as well as the stability of the virus.
5.4 Antigen Presentation During Human SARS-CoV-2 Infection
As an antiviral mechanism, antigen Presenting Cells (APC) are involve in the presentation of viral antigenic peptides in complexed with MHC (major histocompatibility complex) class I and class II molecules to CD8 and CD4 T cells. The selection of peptides and presentation technique of the host leads to a better understanding of cellular immunity and vaccine advancement. During any viral infection, DCs (dendritic cells) play a very important role as an APC. DCs are a linkage between innate and adaptive immunity. Studies deciphering the mechanism of antigen presentation during SARS-CoV-2 infection is not studied well therefore, the mechanism of antigen presentation can be understood based on the available data of predecessor strain infection (SARS-CoV & MERS-CoV) due to its analogy (Chen et al. 2010). Because DCs are found in the respiratory tract and react back whenever there is an inflammation response, DCs are found to be a potential candidate in antigen presentation during SARS infection and also in understanding the immunopathology of SARS (Lau et al. 2012).
During SARS infection, the upregulation of few chemokines such as IP-10 and MP1 is seen very significantly, also few of the antiviral cytokines are found to be low in expression such as IFN-alpha, IFN-beta, and IFN-gamma, and TNF-alpha and IL-6 are found to be at moderate upregulation (Kuri and Weber 2010). Modulation of Toll-like receptors from TLR-1 to TLR-10 was seen to be at the same level; hence no modulation but chemokine receptors such as CCR5, CCR3, and CCR1 are found to be at significant level of upregulation (Law et al. 2009). When similar type of study is conducted in patients infected with Middle East Respiratory Syndrome Coronavirus (MERS-CoV), it has been observed that this virus infects DCs very prolifically by inducing higher expression of IFN-gamma and even cytokines and chemokine related with IFN-gamma are found to be at a higher level. Altogether, antigen presentation in case of MERS-CoV-infected dendritic cells is seen to be significantly higher than in SARS-CoV-infected dendritic cells.
In case of SARS-CoV, the antigen presentation is done most importantly by MHC-I followed by MHC-II (Wieczorek et al. 2017). Studies were done in human macrophages during SARS-CoV infection indicating an interesting observation that severe acute respiratory syndrome coronavirus infects human macrophages due to antibody enhancement (ADE) mediated by IgG. However, macrophages infected with SARS-CoV did not show productive viral replication (Yip et al. 2014). MHC class II and I are further analyzed for epitope mapping which is one of the key steps towards vaccine development and which is studied intensively in case of SARS-CoV-2 infection (Sarkar et al. 2020). Post MHC-I analysis, it was observed that the protein sequence of CoVID-19 is introduced by alleles of HLA-C rather than HLA-A and HLA-B (Prompetchara et al. 2020). Alleles of HLA are studied in case of SARS-CoV infection and have been categorized into protection alleles (HLA-Cw1502, HLA-DR0301, and HLA-A∗0201) and susceptible alleles (HLA-B∗0703, HLA-Cw∗0801, HLA-DR B1∗1202, and HLA-B∗4601) (Wang et al. 2011). In case of MERS-CoV infection, molecules of MHC class II, HLA-DQB1∗02:0, and HLA-DRB1∗11:01 are related with the susceptibility to infection (Fast and Chen 2020; Hajeer et al. 2016). While analyzing the viral genome of COVID-19 for epitope prediction, around 405 T cell epitopes have been determined which exhibits effective affinity towards MHC class I and II in addition two potent neutralizing epitopes (B cell epitope) based on Spike (S) protein were found (Fast and Chen 2020).
5.5 Humoral Immune Response
Humoral immune response is an antibody-mediated immune response. T helper cells assist B cells to differentiate into plasma cells, which in return produces antibodies (Abs) specific to a viral antigen (Ag). In order to limit infection, an antibody which is of neutralizing nature is efficient in fully blocking the virus from entering into host cells and hence plays a very intense protective role at a later stage of infection and also prevents relapse of infection in the future. In case of SARS-CoV, in order to enhance the humoral immune response, both B and T cell epitopes are being studied extensively and mapped for their structural as well as the envelope protein (S, N, M, E) (Channappanavar et al. 2014). During the infection of SARS-CoV-2 it is studied and known that ACE2 (Angiotensin-Converting Enzyme 2) is the receptor present in the host cells and hence the site (B cell epitope site) wherein the Ab-binding site is located is a critical feature to be studied for a valuable insight (Yu et al. 2007). It has been observed that SARS-CoV and 2019-nCoV spike proteins are similar in their major structural and T cell epitope (Kumar et al. 2020).
In order to produce neutralizing Abs, both the proteins need to have B cell epitopes as their receptor binding domains (RBD). A very efficient T cell response has been found to be in relation with the production of higher neutralizing antibodies. T cell epitopes do not require a specific location in contrast to B cell epitopes; hence, it is located anywhere in viral protein. The helper T cell plays a role in isotype switching also and in case of SARS-CoV, the antibody profile of this virus produces IgM and IgG and at a later phase sero-conversion has been observed which is mediated by the helper T cells (Li et al. 2008). IgM disappears at the end of week 12 whereas IgG has been found to last for a longer time pointing out to the probability of IgG being a potent protector Ab during the infection (Li et al. 2003). Current evidence strongly indicates that Th1 type response is key to the successful control of SARS-CoV and MERS-CoV and probably true for SARS-CoV-2 as well (Yong et al. 2019).
5.6 Cellular Immune Response
Cellular immune response is a mechanism of adaptive immunity. Cellular immunity in contrast to the humoral immune response can be seen inside the infected cells, which is mediated by T-lymphocytes. Helper T cells direct the overall adaptive immune response while cytotoxic T cells play a vital role in clearance and killing of viral infected cells. For any effective vaccine advancement, cellular immunity provided by T cells is very much essential as shown by the mouse model experiment on MERS-CoV and SARS-CoV (Yong et al. 2019) wherein their reports suggested that the lack of T cells resulted in no viral clearance in infected mice, hence explaining the importance of T cells in viral infection (Lee et al. 2012).
Referring back to the case of infection caused by Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus, it is reported that CD4+ (TNFα, IL-2, and IFN) and CD8+ (TNFα, IFNγ) memory T cells could persist in SARS-CoV-recovered patient for 4 years and can function by proliferating T cell, producing IFN-gamma, and by DTH response (Kuri and Weber 2010). When investigated 14 out of 23 SARS-recovered patients, post 6 years of infection, it was reported that distinct T cell memory responded to the S library of peptide of SARS-CoV (Channappanavar et al. 2014). Similar findings of distinct CD8+ T cells were seen during the case of MERS-CoV clearance in a mouse model too (Coleman et al. 2016). Hence, this information can be useful in case of SARS-CoV-2 as well. However, in case of SARS-CoV-2 recent reports suggest that the PBMCs of SARS-CoV-2 infected individuals have shown efficient reduction in the CD8+ and CD4+ T cell counts, which may results in compromised T memory cell generation and persistence in SARS-CoV-2 survivors.
5.7 Cytokine Storm Syndrome in Patients Infected with SARS-CoV-2
Cytokine storm syndrome is when there is a fatal blow up of cytokines due to over-reaction by the human body’s immune system in response to an intruder. The Lancet has published a report on cytokine storm syndrome as being one of the causes of CoVID-19 severity (Coleman et al. 2016). This report is backed up by the data on one of the major factors of death due to SARS-CoV-2 infection, which is acute respiratory distress syndrome (ARDS). ARDS has a significant relation with cytokine storm syndrome because, during ARDS, the immune effector cells have been shown to release huge amounts of chemokines and proinflammatory cytokines, which result in a fatal unconfined or uncontrolled systemic inflammatory response (Yao et al. 2020). Previous pandemics caused by coronaviruses such as MERS-CoV and SARS-CoV have also shown such a massive release of chemokines and cytokines: in case of SARS-CoV, CCL2, CCL3, CCl5, CXCL8, CXCL9, CXCL10, etc. and IL-12, IL-18, IL-6, IL-1beta, IL-33, IFN-alpha, IFN-gamma, TNF- alpha & TGF-beta. In case of MERS-CoV, the elevation was seen in the levels of cytokines-IFN-α, IL-6, and chemokine such as CXCL-10, CCL-5, and CXCL-8 (Zheng et al. 2020).
ARDS due to cytokine storm triggers a damaging attack to the body by the immune system causing failure of multiple organs subsequently and leads to death as reported in the case of SARS-CoV-2 outbreak which was the same as the cases of SARS-CoV and MERS-CoV infection reported previously. Lately, drugs targeting IL-18, IL-1, IL-6, and Interferon-gamma have been found effective in treating cytokine storm syndrome in other viral infections for the treatment and therefore may be used for the treatment of the COVID-19 patients for reducing the severity (Cameron et al. 2007). However, one of such drugs falling under the same category, which blocks IL-6, has been reported to be efficient in a few cases of COVID-19 in China (Mehta et al. 2020).
5.8 Immune Evasion Strategies for Coronaviruses
Human CoVs are the one of the most pathogenic viral infections that develops various immune evasion strategies. Studies have come up with reports supporting the fact that the family of CoVs are significantly able to suppress human immune responses by evading the immune detection mode smartly (Kikkert 2020). This immune evasion property might explain longer incubation period, which is of 2–11 days, moderately, if not completely. Immune evasion helps them to efficiently dodge the detection by cellular PRRs of host immune response at the initial phase of infection. The three recent CoVs such as SARS-CoV, MERS-CoV, and SARS-CoV-2 share the same component of immune evasion method since all of them belong to the same genera, Betacoronavirus. The strategy of how these CoVs evades and regulates human immune responses has been a highly talked, studied, and evaluated topic for a very long time. Several studies have been carried out on SARS-CoV and MERS-CoV, which can also be referred in the case of SARS-CoV-2. During SARS-CoV infection, the isolation of viral dsRNA takes place inside the double membrane vesicles (DMVs), which is the probable shield of viral PAMPs from detection by cytosolic PRRs.
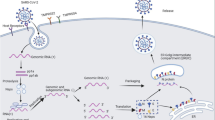
In order to exist and expand inside a host with inbuilt strong antiviral IFN immune responses, CoVs have been seen to employ different levels of strategies (Fig. 5.1) against the innate immune responses, especially type I IFN responses starting with the IFN signaling, induction of IFN, or antiviral action of ISG products (Channappanavar et al. 2016). In any viral infection, interferons play the role of a very potent cytokine in order to control the infection. This suggests that CoVs play its smart game by interfering with the core system of IFN or it can also do so by destroying the key regulators.
5.9 Immune Escape Strategies
5.9.1 Inhibition of IFN Induction
To understand this strategy briefly, both the CoVs, SARS-CoV and MERS-CoV, seem to be less generous regarding inducing IFN into most of the cell types. This less or delayed induction of IFN activates the proinflammatory cytokines and macrophages into the lungs and hence results in leakage of vascular vessels and also impairs the adaptive immune responses (Channappanavar et al. 2016). Exceptional cell type wherein the IFN induction is at a relatively higher level is pDCs, which have shown to be expressing increased levels of Interferon alpha or beta (IFN-alpha & -beta) in case of disease caused by both MERS-CoV and SARS-CoV. This occurs through TLR7. Hence, TLR-like pathway could be a candidate for more core studies in case of SARS-CoV (Totura et al. 2015). In case of MERS-CoV, the protein which inhibits IFN induction is seen to be ORF4a which interacts with dsRNA and the cofactor of RLR, PACT.
5.9.2 Inhibition of IFN Signaling
From IFN docking to ISGF3, STAT1/STAT2/IRF9 complex mediated upregulation of ISGs, SARS-CoV and MERS-CoV act by interfering with the signal transduction chain with the help of several proteins. ORF3a protein and ORF6 protein are reported to decrease the IFNAR levels by proteolytic degradation and ubiquitination and by disrupting STAT1 nuclear import, respectively (Kopecky-Bromberg et al. 2007; Frieman et al. 2007). The ORF4a protein also inhibits IFN induction and hence moderates the ISG expression. The immune evasion strategy also evades adaptive immune responses during infection with SARS-CoV and MERS-CoV wherein T cell activation is diminished by downregulating antigen presentation by MHC I and II molecules.
5.10 Conclusions
The emergence and outbreak caused by SARS-CoV-2 is determined by the disruption of the host immune system by the virus. The virus strain has been observed to disturb the immune system by evasion of the immune response when a person is infected. It is on the safer side to predict that coronaviruses, having the biggest RNA genome so far, do not depend upon sole harmfulness factors; however they utilize a few layers of hostile to IFN procedures such as factors which include the type of virus, the titer of virus, and the load of virus. Else they would not have the strategy to exist, and even expand or mutate to a newer territory with strong antiviral IFN reactions (host). With just their genetic blueprint made available so far, it has become difficult for all research groups to identify the potential molecular targets of the virus. Immunopathology studies of disease caused by SARS-CoV-2 are still being investigated globally. However, with reference to the studies done so far on its predecessor infection, it can be concluded that few of the mechanisms which explain the severity of the disease caused by SARS-CoV-2 is the enzyme Furin, which is found at its activation site, and the mechanism of cytokine storm and immune evasion.
5.11 Future Perspectives
There are many hurdles in designing a vaccine against CoVID-19 due to the fact that different age group and individuals with pre-existing conditions react differently to this disease because of their differences in immune responses, which is why one-size vaccine will not be enough but still research groups are indulged in screening vaccine and the current vaccine candidate used is an antigen of the spike protein of coronavirus. Due to the lack of many failed antiviral strategies in order to efficiently treat infections by coronavirus, scientists are trying to come up with preventive measure such as vaccination. Taking into account the previous cases of coronavirus infection such as the infection of SARS-CoV and MERS-CoV, research groups have managed to come up till the stage of clinical trials of few vaccines and which is an advancement when it comes to the fight against such outbreaks. Hence, it will pave the way for some advancement in designing drugs and vaccine against CoVID-19 too. This is possibly a positive outcome for the vaccine candidate for SARS-CoV-2 disease in the future.
Acknowledgments and Disclosures
The authors are grateful to the Vice Chancellor, King George’s Medical University (KGMU), Lucknow, India, for encouraging this work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, Muller MP, Gold WL, Richardson SE, Poutanen SM, Willey BM, DeVries ME, Fang Y, Seneviratne C, Bosinger SE, Persad D, Wilkinson P, Greller LD, Somogyi R, Humar A, Keshavjee S, Louie M, Loeb MB, Brunton J, McGeer AJ, Canadian SARS Research Network, Kelvin DJ (2007) Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol 81(16):8692–8706
Channappanavar R, Zhao J, Perlman S (2014) T cell-mediated immune response to respiratory coronaviruses. Immunol Res 59(1–3):118–128. https://doi.org/10.1007/s12026-014-8534-z
Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S (2016) Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19(2):181–193
Chen YZ, Liu G, Senju S, Wang Q, Irie A, Haruta M, Matsui M, Yasui F, Kohara M, Nishimura Y (2010) Identification of SARS-COV spike protein-derived and HLA-A2-restricted human CTL epitopes by using a new muramyl dipeptide derivative adjuvant. Int J Immunopathol Pharmacol 23(1):165–177
Coleman CM, Sisk JM, Halasz G, Zhong J, Beck SE, Matthews KL, Venkataraman T, Rajagopalan S, Kyratsous CA, Frieman MB (2016) CD8+ T cells and macrophages regulate pathogenesis in a mouse model of Middle East respiratory syndrome. J Virol. 91(1). pii: e01825-16. https://doi.org/10.1128/JVI.01825-16
Drosten C, Günther S, Preiser W et al (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348(20):1967–1976. https://doi.org/10.1056/NEJMoa030747
Fast E, Chen B (2020) Potential T-cell and B-cell epitopes of 2019-nCoV. bioRxiv 2020.02.19.955484. https://doi.org/10.1101/2020.02.19.955484
Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282:1–23. https://doi.org/10.1007/978-1-4939-2438-7_1
Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS (2007) Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81(18):9812–9824
Hajeer AH, Balkhy H, Johani S, Yousef MZ, Arabi Y (2016) Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Ann Thorac Med 11(3):211–213. https://doi.org/10.4103/1817-1737.185756
Khan S, Siddique R, Shereen MA, Ali A, Liu J, Bai Q, Bashir N, Xue M (2020) The emergence of a novel coronavirus (SARS-CoV-2), their biology and therapeutic options. J Clin Microbiol. pii: JCM.00187-20. https://doi.org/10.1128/JCM.00187-20
Kikkert M (2020) Innate immune evasion by human respiratory RNA viruses. J Innate Immun 12(1):4–20. https://doi.org/10.1159/000503030
Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P (2007) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81(2):548–557
Kumar S, Maurya VK, Prasad AK et al (2020) Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). VirusDis 31:13–21. https://doi.org/10.1007/s13337-020-00571-5
Kuri T, Weber F (2010) Interferon interplay helps tissue cells to cope with SARS-coronavirus infection. Virulence 1(4):273–275. https://doi.org/10.4161/viru.1.4.11465
Lau YL, Peiris JS, Law HK (2012) Role of dendritic cells in SARS coronavirus infection. Hong Kong Med J 18(Suppl 3):28–30
Law HK, Cheung CY, Sia SF, Chan YO, Peiris JS, Lau YL (2009) Toll-like receptors, chemokine receptors and death receptor ligands responses in SARS coronavirus infected human monocyte derived dendritic cells. BMC Immunol 10:35. https://doi.org/10.1186/1471-2172-10-35
Lee S, Stokes KL, Currier MG, Sakamoto K, Lukacs NW, Celis E, Moore ML (2012) Vaccine-elicited CD8+ T cells protect against respiratory syncytial virus strain A2-line19F-induced pathogenesis in BALB/c mice. J Virol 86(23):13016–13024. https://doi.org/10.1128/JVI.01770-12
Li F (2016) Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3(1):237–261
Li G, Chen X, Xu A (2003) Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med 349(5):508–509
Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, Tang X, Temperton NJ, Weiss RA, Brenchley JM, Douek DC, Mongkolsapaya J, Tran BH, Lin CL, Screaton GR, Hou JL, McMichael AJ, Xu XN (2008) T cell responses to whole SARS coronavirus in humans. J Immunol 181(8):5490–5500
Li X, Zai J, Zhao Q et al (2020) Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol 2020:1–10. https://doi.org/10.1002/jmv.25731. [Published online ahead of print, 2020 Feb 27]
Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ; HLH Across Speciality Collaboration, UK (2020) COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395(10229):1033–1034. https://doi.org/10.1016/S0140-6736(20)30628-0
Prompetchara E, Ketloy C, Palaga T (2020) Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol 38(1):1–9. https://doi.org/10.12932/AP-200220-0772
Sarkar B, Ullah MA, Johora FT, Taniya MA, Araf Y (2020) The essential facts of Wuhan novel coronavirus outbreak in China and epitope-based vaccine designing against COVID-19. bioRxiv 2020.02.05.935072. https://doi.org/10.1101/2020.02.05.935072
Totura AL, Whitmore A, Agnihothram S, Schäfer A, Katze MG, Heise MT, Baric RS (2015) Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio 6(3):e00638–e00615. https://doi.org/10.1128/mBio.00638-15
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, Function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 180:1–12. https://doi.org/10.1016/j.cell.2020.02.058. [Epub ahead of print]
Wang SF, Chen KH, Chen M, Li WY, Chen YJ, Tsao CH, Yen MY, Huang JC, Chen YM (2011) Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol 24(5):421–426. https://doi.org/10.1089/vim.2011.0024
Wang Y, Wang Y, Chen Y, Qin Q (2020) Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol 2020:1–9. https://doi.org/10.1002/jmv.25748. [Epub ahead of print]
Wieczorek M, Abualrous ET, Sticht J, Álvaro-Benito M, Stolzenberg S, Noé F, Freund C (2017) Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front Immunol 8:292. https://doi.org/10.3389/fimmu.2017.00292
World Health Organization (2020) Coronavirus disease 2019 (COVID-19) situation report—51. World Health Organization. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10. Accessed 16 Mar 2020
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. pii: eabb2762. https://doi.org/10.1126/science.abb2762
Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, Zhan S, Lu R, Li H, Tan W, Liu D (2020) In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. pii: ciaa237. https://doi.org/10.1093/cid/ciaa237
Yip MS, Leung NH, Cheung CY, Li PH, Lee HH, Daëron M, Peiris JS, Bruzzone R, Jaume M (2014) Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J 11:82. https://doi.org/10.1186/1743-422X-11-82
Yong CY, Ong HK, Yeap SK, Ho KL, Tan WS (2019) Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front Microbiol 10:1781. https://doi.org/10.3389/fmicb.2019.01781
Yu H, Jiang LF, Fang DY, Yan HJ, Zhou JJ, Zhou JM, Liang Y, Gao Y, Zhao W, Long BG (2007) Selection of SARS-coronavirus-specific B cell epitopes by phage peptide library screening and evaluation of the immunological effect of epitope-based peptides on mice. Virology 359(2):264–274
Zheng H, Zhang M, Yang C et al (2020) Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol, 1–3. https://doi.org/10.1038/s41423-020-0401-3
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, S., Nyodu, R., Maurya, V.K., Saxena, S.K. (2020). Host Immune Response and Immunobiology of Human SARS-CoV-2 Infection. In: Saxena, S. (eds) Coronavirus Disease 2019 (COVID-19). Medical Virology: From Pathogenesis to Disease Control. Springer, Singapore. https://doi.org/10.1007/978-981-15-4814-7_5
Download citation
DOI: https://doi.org/10.1007/978-981-15-4814-7_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4813-0
Online ISBN: 978-981-15-4814-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)