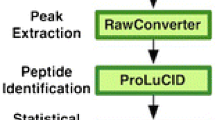
Abstract
Proteomics, a field of bioinformatics majorly deals with the study of proteins and its structures in a predefined set of conditions. Integration of this field of bioinformatics with mathematical approaches like statistics, machine learning techniques, and various data-scaling methods not only fetches new discoveries in this field but also offers results with great accuracy and precision. This chapter dives its readers into the scope of machine learning algorithms in the study of proteins in a more extensive manner, provides examples of various real-time datasets that can be used to analyze the proteins, explains many preprocessing techniques that could be applied to these datasets for dimension reduction of the dataset, and briefs about the machine learning algorithms that are widely used along with the applications and comparison of these algorithms in terms of its performance and usage. This article also supplements with two case studies which revolve about the application of an algorithm in a real-world datasets.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Can T (2013) Introduction to bioinformatics. Part of the Methods in Molecular Biology book series (MIMB, vol 1107), pp 51–71. https://doi.org/10.1007/978-1-62703-748-8_4
Lesk AM (2019) Bioinformatics. https://www.britannica.com/science/bioinformatics
Yee A, Pardee K, Christendat D, Savchenko A, Edwards AM, Arrowsmith CH (2003) Structural proteomics: toward high-throughput structural biology as a tool in functional genomics. https://doi.org/10.1021/ar010126g
Introduction to Proteomics, Wikibooks. https://en.wikibooks.org/wiki/Proteomics/Introduction_to_Proteomics, 2017
Center for Proteomics and Bioinformatics, Expression Proteomics, Western Reserve University, Cleveland, Ohio. http://proteomics.case.edu/proteomics/expression-proteomics.html, 2010
Center for Proteomics and Bioinformatics, Interaction Proteomics, Western Reserve University, Cleveland, Ohio. http://proteomics.case.edu/proteomics/interaction-proteomics.html, 2010
Yokota H (2019) Applications of proteomics in pharmaceutical research and development. Appl Proteomics Pharm Res Dev
Larranaga P, Calvo B, Santana R, Bielza C, Galdiano J, Inza I, Lozano JA, Armananzas R, Santafe G, Perez A, Robles V (2005) Machine learning in bioinformatics. Brief Bioinform
Artificial intelligence boosts proteome research, Technical University of Munich (TUM). https://www.sciencedaily.com/releases/2019/05/190529113044.htm, 2019
Strimbu K, Tavel JA (2010) What is biomarker? Curr Opin HIV AIDS 5(6):463–466
Swan AL, Mobasheri A, Allaway D, Liddell S, Bacardit J (2013) Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology. OMICS: J Integr Biol
Fan Z, Kong F, Zhou Y, Chen Y, Dai Y (2018) Intelligence algorithms for protein classification by mass spectrometry. BioMed Res Int
Sampson DL, Parker TJ, Upton Z, Hurst CP (2011) A comparison of methods for classifying clinical samples based on proteomics data: a case study for statistical and machine learning approaches. PLoS ONE 6(9):e24973. https://doi.org/10.1371/journal.pone.0024973
Dimensionality reduction, Wikipedia. https://en.wikipedia.org/wiki/Dimensionality_reduction, 2016
Sharma P (2018) The ultimate guide to 12 dimensionality reduction techniques (with Python codes). https://www.analyticsvidhya.com/blog/2018/08/dimensionality-reduction-techniques-python/
Naveenkumar KS, Mohammed Harun Babu R, Vinayakumar R, Soman KP (2018) Protein family classification using deep learning. Center for Computational Engineering and Networking (CEN)
Geurts P, Fillet M, de Seny D, Meuwis M-A, Malaise M, Merville M-P, Wehenkel L (2005) Proteomic mass spectra classification using decision tree based ensemble methods. Oxford Academic
He B, Zhang B (2013) Discovery of proteomics based on machine learning. Beihang University
Liu Q, Qiao M, Sung AH (2008) Distance metric learning and support vector machines for classification of mass spectrometry proteomics data. In: Seventh international conference on machine learning and applications
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kiranmai, V.P., Siddesh, G.M., Manisekhar, S.R. (2020). Supervised Techniques in Proteomics. In: Srinivasa, K., Siddesh, G., Manisekhar, S. (eds) Statistical Modelling and Machine Learning Principles for Bioinformatics Techniques, Tools, and Applications. Algorithms for Intelligent Systems. Springer, Singapore. https://doi.org/10.1007/978-981-15-2445-5_12
Download citation
DOI: https://doi.org/10.1007/978-981-15-2445-5_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-2444-8
Online ISBN: 978-981-15-2445-5
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)