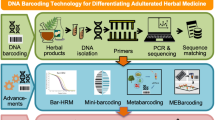
Abstract
Correctly identified medicinal materials are the premise of research and applications on Chinese medicine. DNA authentication is an accurate method for routine identification of medicinal herbs. The molecular authentication procedure involving DNA sequencing takes five steps: (a) to extract DNA from the sample, (b) to amplify a specific region of the genome by polymerase chain reaction; (c) to sequence the PCR product; (d) to find the available DNA sequences in a nucleotide database; and (e) to compare the unknown sequence with the available reference sequences. Molecular authentication can also be done by DNA fingerprinting that assess the whole or specific region of genome. In the process, informatics assists in: (a) providing a means for storing, sorting, retrieving, and analyzing of experimental data; (b) providing computer programs for identifying the unknown sample by matching with the reference sequences; and (c) aiding the design of appropriate testing procedures and identification tools. This chapter introduces several public databases for the search of taxonomic and nucleotide information, such as Flora of China, NCBI Nucleotide database, Barcode of Life Data Systems (BOLD), and Medicinal Materials DNA Barcode Database (MMDBD). The procedures of mass retrieving and sorting of sequences from databases will be demonstrated. The pros and cons of various DNA sequence formats like FASTA, Abstract Syntax Notation 1 (ASN1), eXtensible Markup Language (XML) will be discussed. Finally, the precautions of using software packages for performing sequence alignment, matching species in databases, and constructing phylogenetic trees will be described.
Ka-Lok Wong and Yat-Tung Lo: Equal contribution.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Luscombe NM, Greenbaum D, Gerstein M. What is bioinformatics? A proposed definition and overview of the field. Methods Inf Med. 2001;40(4):346–58.
Hogeweg P, Hesper B. Interactive instruction on population interactions. Comput Biol Med. 1978;8(4):319–27.
Hogeweg P. The roots of bioinformatics in theoretical biology. PLoS Comput Biol. 2011;7(3):e1002021.
Watson JD, Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–8.
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269(5223):496–512.
State Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China: part 1. Beijing: China Medical Science Press; 2010.
Editorial Team. National Center for Biotechnology Information. Published on the Internet http://www.ncbi.nlm.nih.gov. Accessed 1 June 2015.
Lou SK, Wong KL, Li M, But PP, Tsui SK, Shaw PC. An integrated web medicinal materials DNA database: MMDBD (Medicinal Materials DNA Barcode Database). BMC Genom. 2010;11:402.
Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proceedings Biological Sciences/The Royal Society. 2003;270(1512):313–21.
Zhang GJ. Methodology for Chinese medicine authentication research. Beijing: People’s Medical Publishing House; 2010.
Swango KL, Timken MD, Chong MD, Buoncristiani MR. A quantitative PCR assay for the assessment of DNA degradation in forensic samples. Forensic Sci Int. 2006;158(1):14–26.
Radstrom P, Knutsson R, Wolffs P, Lovenklev M, Lofstrom C. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol Biotechnol. 2004;26(2):133–46.
Dellaporta S, Wood J, Hicks J. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1(4):19–21.
Kang H, Cho Y, Yoon U, Eun M. A rapid DNA extraction method for RFLP and PCR analysis from a single dry seed. Plant Mol Biol Rep. 1998;16(1):90.
Draper J, Scott R. The isolation of plant nucleic acids. In: Draper J, Scott R, Armitage P, Walden R, editors. Plant genetic transformation and gene expression. A Laboratory Manual, Blackwell, London; 1988. p. 199–236.
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjoller R, Koljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. Fungal community analysis by high-throughput sequencing of amplified markers—a user’s guide. New Phytol. 2013;199(1):288–99.
Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001.
CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106(31):12794–7.
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6.
Ye HG, Zou B. Chinese medicinal plants. Beijing: Chemical Industry Press; 2014.
eFloras. Flora of China. Missouri Botanical Garden, St. Louis, MO & Harvard University Herbaria, Cambridge, MA. Published on the Internet http://www.efloras.org (2008). Accessed 1 June 2015.
European Bioinformatics Institute. EMBL Nucleotide Sequence Database. Published on the Internet http://www.embl.org. Accessed 1 June 2015.
Center for Information Biology and DNA Data Bank of Japan. DNA Data Bank of Japan. Published on the Internet http://www.ddbj.nig.ac.jp. Accessed 1 June 2015.
Ratnasingham S, Hebert PD. Bold: the barcode of life data system. Mol Ecol Notes. 2007;7(3):355–64. http://www.barcodinglife.org.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;45:95–8.
W3C. XML technology. Published on the Internet http://www.w3.org/standards/xml. Accessed 1 June 2015.
Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73(1):237–44.
Li M, Wong K, Chan W, Li J, But P-H, Cao H, Shaw P-C. Establishment of DNA barcodes for the identification of the botanical sources of the Chinese ‘cooling’ beverage. Food Control. 2012;25(2):758–66.
European Bioinformatics Institute. ClustalW help file. Published on the Internet http://www.ebi.ac.uk/Tools/msa/clustalw2/help/faq.html#21. Accessed 1 June 2015.
Ha WY, Reid DG, Kam WL, Lau YY, Sham WC, Tam SY, Sin DW, Mok CS. Genetic differentiation between fake abalone and genuine Haliotis species using the forensically informative nucleotide sequencing (FINS) method. J Agric Food Chem. 2011;59(10):5195–203. doi:10.1021/jf104892n.
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA. 2005;102(23):8369–74.
Guo X, Wang X, Su W, Zhang G, Zhou R. DNA barcodes for discriminating the medicinal plant Scutellaria baicalensis (Lamiaceae) and its adulterants. Biol Pharm Bull. 2011;34(8):1198–203.
Liu Z, Chen KL, Luo K, Pan HL, Chen SL. DNA barcoding in medicinal plants Caprifoliaceae. China J Chin Materia Medica. 2010;35(19):3527.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531–5.
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23(21):4407–14.
PREMIER Biosoft. PCR primer design guidelines. Published on the Internet http://www.premierbiosoft.com/tech_notes/PCR_Primer_Design.html. Accessed 1 June 2015.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115.
Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–91.
Coghlan ML, Haile J, Houston J, Murray DC, White NE, Moolhuijzen P, Bellgard MI, Bunce M. Deep sequencing of plant and animal DNA contained within traditional Chinese medicines reveals legality issues and health safety concerns. PLoS Genet. 2012;8(4):e1002657.
Cheung VG, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Making and reading microarrays. Nat Genet. 1999;21(1 Suppl):15–9.
Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21(1 Suppl):10–4.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media Singapore
About this chapter
Cite this chapter
Wong, KL., Lo, YT., Shaw, PC. (2016). Bioinformatics for Molecular Authentication of Chinese Medicinal Materials. In: Leung, Sw., Hu, H. (eds) Evidence-based Research Methods for Chinese Medicine. Springer, Singapore. https://doi.org/10.1007/978-981-10-2290-6_8
Download citation
DOI: https://doi.org/10.1007/978-981-10-2290-6_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-2289-0
Online ISBN: 978-981-10-2290-6
eBook Packages: MedicineMedicine (R0)