Abstract
The classification of benign versus malignant lung nodules using chest CT plays a pivotal role in the early detection of lung cancer and this early detection has the best chance of cure. Although deep learning is now the most successful solution for image classification problems, it requires a myriad number of training data, which are not usually readily available for most routine medical imaging applications. In this paper, we propose the transferable multi-model ensemble (TMME) algorithm to separate malignant from benign lung nodules using limited chest CT data. This algorithm transfers the image representation abilities of three ResNet-50 models, which were pre-trained on the ImageNet database, to characterize the overall appearance, heterogeneity of voxel values and heterogeneity of shape of lung nodules, respectively, and jointly utilizes them to classify lung nodules with an adaptive weighting scheme learned during the error back propagation. Experimental results on the benchmark LIDC-IDRI dataset show that our proposed TMME algorithm achieves a lung nodule classification accuracy of 93.40%, which is markedly higher than the accuracy of seven state-of-the-art approaches.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The 2015 global cancer statistics show that lung cancer accounts for approximately 13% of 14.1 million new cancer cases and 19.5% of cancer-related deaths each year [1]. The 5-year survival for patients with an early diagnosis is approximately 54%, as compared to 4% if the diagnosis is made late when the patient has the stage IV disease [2]. Hence, early diagnosis and treatment are the most effective means to improve lung cancer survival. The National Lung Screening Trial [3] showed that screening with CT will result in a 20% reduction in lung cancer deaths. On chest CT scans, a “spot” on the lung, less than 3 cm in diameter, is defined as a lung nodule, which can be benign or malignant [4]. Malignant nodules may be primary lung tumors or metastases and so the classification of lung nodules is critical for best patient care.
Radiologists typically read chest CT scans on a slice-by-slice basis, which is time-consuming, expensive and prone to operator bias and requires a high degree of skill and concentration. Computer-aided lung nodule classification avoids many of these issues and has attracted a lot of research attention. Most solutions in the literature are based on using hand-crafted image features to train a classifier, such as the support vector machine (SVM), artificial neural network and so on. For instance, Han et al. [5] extracted Haralick and Gabor features and local binary patterns to train a SVM for lung nodule classification. Dhara et al. [6], meanwhile, used computed shape-based, margin-based and texture-based features for the same purpose.
More recently, deep learning, particularly the deep convolutional neural network (DCNN), has become the most successful image classification technique and it provides a unified framework for joint feature extraction and classification [7]. Hua et al. applied the DCNN and deep belief network (DBN) to separate benign from malignant lung nodules. Shen et al. [7] proposed a multi-crop convolutional neural network (MC-CNN) for lung nodule classification. Despite improved accuracy, these deep models have not achieved the same performance on routine lung nodule classification as they have in the famous ImageNet Challenge. The suboptimal performance is attributed mainly to the overfitting of deep models due to inadequate training data, as there is usually a small dataset in medical image analysis and this relates to the work required in acquiring the image data and then in image annotation. Hu et al. [8] proposed a deep transfer metric learning method to transfer discriminative knowledge from a labeled source domain to an unlabeled target domain to overcome this limitation.
A major difference between traditional and deep learning methods is that traditional methods rely more on the domain knowledge, such as there is a high correspondence between nodule malignancy and heterogeneity in voxel values (HVV) and heterogeneity in shapes (HS) [9], and deep learning relies on access to massive datasets. Ideally, the advantages of both should be employed. Chen et al. [10] fused heterogeneous Haralick features, histogram of oriented gradient (HOG) and features derived from the deep stacked denoising autoencoder and DCNN at the decision level to predict nine semantic labels of lung nodules. In our previous work [11], we used a texture descriptor and a shape descriptor to explore the heterogeneity of nodules in voxel values and shape, respectively, and combined both descriptors with the features learned by a nine-layer DCNN for nodule classification. Although improved accuracy was reported, this method still uses hand-crafted features to characterize the heterogeneity of nodules, which are less effective. Recently, Hu et al. [8] reported that the image representation ability of DCNNs, which was learned from large-scale datasets, could be transferred to solving generic small-data visual recognition problems. Hence, we suggest transferring the DCNN’s image representation ability to characterize the overall appearance of lung nodule images and also the nodule heterogeneity in terms of voxel values and shape.
In this paper, we propose a transferable multi-model ensemble (TMME) algorithm for benign-malignant lung nodule classification on chest CT. The main uniqueness of this algorithm includes: (1) three types of image patches are designed to fine-tune three pre-trained ResNet-50 models, aiming to characterize the overall appearance (OA), HVV and HS of each nodule slice, respectively; and (2) these three ResNet-50 models are used jointly to classify nodules with an adaptive weighting scheme learned during the error back propagation, which enables our model to be trained in an ‘end to end’ manner. We compared our algorithm to seven state-of-the-art lung nodule classification approaches on the benchmark LIDC-IDRI dataset. Our results suggest that the proposed algorithm provides substantial performance improvement.
2 Data and Materials
The benchmark LIDC-IDRC databases [12] were used for this study, in which the nodules were evaluated over five levels, from benign to malignant, by up to four experienced thoracic radiologists. The mode of levels given by radiologists was defined as the composite level of malignancy. Same to [5,6,7, 13,14,15], we only considered nodules ≥3 mm in diameter and treated 873 nodules with composite level of 1 or 2 as benign and 484 nodules with composite level of 4 or 5 as malignant.
3 Algorithm
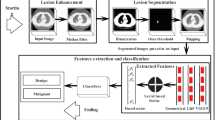
We have summarized our proposed TMME algorithm in Fig. 1. The algorithm has three steps: (1) extracting the region of interest (ROI) for preprocessing and data augmentation, (2) building a TMME model for slice-based nodule classification, and (3) classifying each nodule based on the labels of its slices.
3.1 Preprocessing and Data Augmentation
A lung nodule is presented in multiple slices. On each slice, a square ROI encapsulating the nodule is identified using the tool developed by [16] to represent the nodule’s OA. To characterize the nodule’s HVV, non-nodule voxels outside the ROI are set to 0 and, if the ROI is larger than 16 × 16, a 16 × 16 patch that contains the maximum nodule voxels is extracted. To describe the nodule’s HS, nodule voxels inside the ROI are set to 255. Then, the OA patch, HVV patch and HS patch are resized to 200 × 200, using the bicubic interpolation. Four augmented copies of each training sample are generated by using rotation, shear, horizontal or vertical flip and translation with random parameters to enlarge the size of the training set and are put in the enlarged training set.
3.2 TMME for Nodule Slice Classification
The ResNet-50 model [17] that has been pre-trained on the ImageNet dataset, is adopted. Two neurons in the last fully connected layer are randomly selected and other neurons, together with the weights attached to them, are removed. Three copies of this ResNet-50 are fine-tuned using all OA, HVV and HS patches in the enlarged training set, respectively, to adapt them to characterizing nodule slices. Denoting the prediction vector produced by each ResNet-50 by \( \varvec{X}_{i} = \left( {x_{i1} ,x_{i2} } \right)\,\left( {i = 1,2,3} \right) \), the ultimate prediction vector of the ensemble model can be calculated as:
where \( P_{k} \) is the predicted likelihood of the input belonging to category \( k \), and \( \omega_{ijk} \) is the weight which connects the \( x_{ij} \) and \( P_{k} \). Thus, the integrated loss of this ensemble model can be formulated as:
where \( y\, \in \,\left\{ {1,2} \right\} \) is the input’s true label, and \( \varvec{P} = \left( {P_{1} ,P_{2} } \right) \). The change of weight \( \omega_{ijk} \) in the ensemble model is in proportion to descend along the gradient, shown as follows:
where \( \eta \) represents the learning rate, and, if \( k = y,\delta_{ky} = 1 \), otherwise, \( \delta_{ky} = 0 \).
Since our training data set is small, the learning rate is set to 0.00001 and the stochastic gradient descent with a batch size of 100 is adopted. Moreover, 10% of the training patches are chosen to form a validation set, and the training is terminated even before reaching the maximum iteration number of 50, if the error on the other 90% of training images continues to decline but the error on the validation set stops decreasing.
3.3 Nodule Classification
Let a lung nodule \( \varvec{\varPsi} \) be contained in \( S \) slices, denoted by \( \varvec{\varPsi}= \left\{ {\varPsi_{1} ,\varPsi_{2} , \ldots ,\Psi _{S} } \right\} \). Input the \( i \)-th slice \( \varPsi_{i} \) into the TMME model, and we obtain a two-dimensional prediction vector \( \varvec{H}\left( {\varPsi_{i} } \right) \). The class label of nodule \( \varvec{\varPsi} \) is assigned based on the sum of the prediction made on each slice, shown as follows:

4 Results
The proposed TMME algorithm was applied to the LIDC-IDRC dataset 10 times independently, with 10-fold cross validation. The mean and standard deviation of obtained accuracy, sensitivity, specificity and area under the receiver operator curve (AUC), together with the performance of seven state-of-the-art methods on this dataset, were given in Table 1. It shows that our algorithm not only outperformed hand-crafted feature-based traditional methods but also substantially improved upon Xie et al.’s method [11]. Our results indicate that the pre-trained and fine-tuned ResNet-50 model can effectively transfer the image representation ability learned on the ImageNet dataset to characterizing the OA, HVV and HS of lung nodules, and an adaptive ensemble of these three models has superior ability to differentiate malignant from benign lung nodules.
5 Discussion
5.1 Data Argumentation
The number of training samples generated by data augmentation plays an important role in applying a deep model to small-sample learning problems. On one hand, training a deep model requires as many data as possible; on the other hand, more data always lead to higher time cost. We re-performed the experiment using different numbers of augmented images and listed the obtained performance in Table 2.
It reveals that using four augmented images for each training sample achieved a trade-off between accuracy and time cost, since further increasing the number of augmented images only improved the accuracy slightly but cost much more time for training. Meanwhile, it should be noted that our algorithm, without using data augmentation, achieved an accuracy of 89.84%, which is still superior to the accuracy of those methods given in Table 1.
5.2 Ensemble Learning
To demonstrate the performance improvement that results from the adaptive ensemble of three ResNet-50 models, we compared the performance of our algorithm to that of three component models, which characterize lung nodules from the perspective of OA, HVV and HS, respectively. As shown in Table 3, although each ResNet-50 model achieves a relatively good performance, an adaptive ensemble of them brings a further performance gain.
5.3 Other Pre-trained DCNN Models
Besides ResNet-50, GoogLeNet [18] and VGGNet [19] are two of the most successful DCNN models. Using each of those three models to characterize lung nodules from each of three perspectives, i.e. OA, HVV and HS, we have 27 different configurations. To evaluate the performance of using other DCNN models, we tested all 27 configurations and gave the accuracy and AUC of the top five configurations in Table 4. It shows that ResNet-50 is very powerful and using three ResNet-50 results in the highest accuracy and AUC. Nevertheless, it also suggests that GoogLeNet and VGGNet are good choices as well and using them to replace ResNet-50 may produce very similar accuracy in some configurations.
5.4 Hybrid Ensemble of 27 TMME Models
Using all possible combination of VGGNet, GoogLeNet and ResNet-50 to characterize the OA, HVV and HS of lung nodules, we can have totally 27 proposed TMME models, which can be further combined by using an adaptive weighting scheme learned in the same way. Table 5 shows that the ensemble of 27 TMME models can only slightly improve the classification accuracy, but with a major increase in the computational complexity of training the model.
5.5 Computational Complexity
In our experiments, it took about 12 h to train the proposed model and less than 0.5 s to classify each lung nodule (Intel Xeon E5-2678 V3 2.50 GHz \( \times \) 2, NVIDIA Tesla K40c GPU \( \times \) 2, 128 GB RAM, 120 GB SSD and Matlab 2016). It suggests that the proposed algorithm, though computation very complex during the training process that can be performed offline, is very efficient for online testing and could be used in a routine clinical workflow.
6 Conclusion
We propose the TMME algorithm for benign-malignant lung nodule classification on chest CT. We used three pre-trained and fine-tuned ResNet-50 models to characterize the OA, HVV and HS of lung nodules, and combined these models using an adaptive weighting scheme learned during the back-propagation process. Our results on the benchmark LIDC-IDRC dataset suggest that our algorithm produces more accurate results than seven state-of-the-art approaches.
References
Siegel, R.L., Miller, K.D., Jemal, A.: Cancer statistics. CA Cancer J. Clin. 65(1), 5–29 (2015)
Bach, P.B., Mirkin, J.N., Oliver, T.K., Azzoli, C.G., Berry, D.A., Brawley, O.W., Byers, T., Colditz, G.A., Gould, M.K., Jett, J.R.: Benefits and harms of CT screening for lung cancer: a systematic review. JAMA, J. Am. Med. Assoc. 307, 2418–2429 (2012)
Abraham, J.: Reduced lung-cancer mortality with low-dose computed tomographic screening. New Engl. J. Med. 365, 395–409 (2011)
American Thoracic Society: What is a lung nodule? Am. J. Respir. Crit. Care Med. 193, 11–12 (2016)
Han, F., Wang, H., Zhang, G., Han, H., Song, B., Li, L., Moore, W., Lu, H., Zhao, H., Liang, Z.: Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J. Digit. Imaging 28(1), 99–115 (2015)
Dhara, A.K., Mukhopadhyay, S., Dutta, A., Garg, M., Khandelwal, N.: A combination of shape and texture features for classification of pulmonary nodules in lung CT images. J. Digit. Imaging 29(4), 466–475 (2016)
Shen, W., Zhou, M., Yang, F., Yu, D., Dong, D., Yang, C., Zang, Y., Tian, J.: Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recogn. 61, 663–673 (2017)
Hu, J., Lu, J., Tan, Y.P.: Deep transfer metric learning. In: CVPR 2015, pp. 325–333. IEEE Press, New York (2015)
Metz, S., Ganter, C., Lorenzen, S., Marwick, S.V., Holzapfel, K., Herrmann, K., Rummeny, E.J., Wester, H.J., Schwaiger, M., Nekolla, S.G.: Multiparametric MR and PET imaging of intratumoral biological heterogeneity in patients with metastatic lung cancer using voxel-by-voxel analysis. PLoS ONE 10(7), e0132386 (2014)
Chen, S., Qin, J., Ji, X., Lei, B., Wang, T., Ni, D., Cheng, J.Z.: Automatic scoring of multiple semantic attributes with multi-task feature leverage: a study on pulmonary nodules in CT images. IEEE Trans. Med. Imaging 99, 1 (2016)
Xie, Y., Zhang, J., Liu, S., Cai, W., Xia, Y.: Lung nodule classification by jointly using visual descriptors and deep features. In: Müller, H., et al. (eds.) MCV 2016, BAMBI 2016. LNCS, vol. 10081. Springer, Cham (2017). doi:10.1007/978-3-319-61188-4_11
Iii, S.G.A., Mclennan, G., Bidaut, L., Mcnittgray, M.F., Meyer, C.R., Reeves, A.P., Zhao, B., Aberle, D.R., Henschke, C.I., Hoffman, E.A.: The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 38, 915–931 (2011)
Hua, K.L., Hsu, C.H., Hidayati, S.C., Cheng, W.H., Chen, Y.J.: Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther. 8, 2015–2022 (2015)
Han, F., Zhang, G., Wang, H., Song, B.: A texture feature analysis for diagnosis of pulmonary nodules using LIDC-IDRI database. In: ICMIPE 2013, pp. 14–18. IEEE Press (2013)
Anand, S.K.V.: Segmentation coupled textural feature classification for lung tumor prediction. In: 2010 IEEE International Conference on Communication Control and Computing Technologies, pp. 518–524. IEEE Press, New York (2010)
Lampert, T.A., Stumpf, A., Gançarski, P.: An empirical study into annotator agreement, ground truth estimation, and algorithm evaluation. IEEE TIP 25(6), 2557–2572 (2016)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR 2016, pp. 770–778. IEEE Press, New York (2016)
Szegedy, C., Liu, W., Jia, Y., Sermanet, P.: Going deeper with convolutions. In: CVPR 2015, pp. 1–9. IEEE Press, New York (2016)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409-1556 (2014)
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China under Grants 61471297, in part by the Seed Foundation of Innovation and Creation for Graduate Students in Northwestern Polytechnical University under Grants Z2017041, and in part by the Australian Research Council (ARC) Grants. We acknowledged the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Xie, Y., Xia, Y., Zhang, J., Feng, D.D., Fulham, M., Cai, W. (2017). Transferable Multi-model Ensemble for Benign-Malignant Lung Nodule Classification on Chest CT. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10435. Springer, Cham. https://doi.org/10.1007/978-3-319-66179-7_75
Download citation
DOI: https://doi.org/10.1007/978-3-319-66179-7_75
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66178-0
Online ISBN: 978-3-319-66179-7
eBook Packages: Computer ScienceComputer Science (R0)