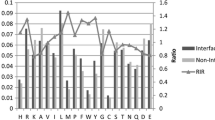
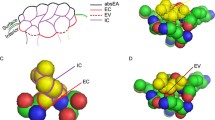
Abstract
Although many studies about near native protein-protein interface recognition have been done in the past thirty years, the formation mechanism of protein-protein interface is still ambiguous. Here, we propose a new probability way to understand protein-protein interface formation mechanism at amino acid level. The probability of two surface residues from different monomers as a true interface residue pair in the complex is estimated by their geometric and physicochemical properties in the structures of protein monomers. The residue pairs with different probabilities combine together to form a protein-protein interface. The probabilities of residue pairs on candidate interfaces are integrated for near native interface recognition. Five simple probability based discriminants are constructed based on the distances and contact areas between residues. The performances are comparable to the ones of the sophisticated methods developed previously. The idea proposed in this work will make positive influence on the future study of protein-protein interactions.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Xue, L.C., Dobbs, D., Bonvin, A.M., Honavar, V.: Computational prediction of protein interfaces: a review of data driven methods. FEBS Lett. 589(23), 3516–3526 (2015). doi:10.1016/j.febslet.2015.10.003
Fischer, E.: Ueber die optischen Isomeren des Traubenzuckers, der Gluconsäure und der Zuckersäure. Berichte der deutschen chemischen Gesellschaft 23(2), 2611–2624 (1890). doi:10.1002/cber.189002302157
Fischer, E.: Einfluss der configuration auf die Wirkung der Enzyme. Berichte der deutschen chemischen Gesellschaft 27(3), 2985–2993 (1894). doi:10.1002/cber.18940270364
Koshland, D.E.: Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 44(2), 98–104 (1958). doi:dx.doi.org/10.1073/pnas.44.2.98
Moont, G., Gabb, H.A., Sternberg, M.J.: Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins 35(3), 364–373 (1999). doi:10.1002/(SICI)1097-0134(19990515)35:3<364::AID-PROT11>3.0.CO;2-4
Vreven, T., Moal, I.H., Vangone, A., Pierce, B.G., Kastritis, P.L., Torchala, M., Chaleil, R., Jimenez-Garcia, B., Bates, P.A., Fernandez-Recio, J., Bonvin, A.M., Weng, Z.: Updates to the integrated protein-protein interaction benchmarks: docking benchmark version 5 and affinity benchmark version 2. J. Mol. Biol. 427(19), 3031–3041 (2015). doi:10.1016/j.jmb.2015.07.016
Kyte, J., Doolittle, R.F.: A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157(1), 105–132 (1982). doi:10.1016/0022-2836(82)90515-0
Eisenberg, D.: Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 53, 595–623 (1984). doi:10.1146/annurev.bi.53.070184.003115
Olsson, M.H., Sondergaard, C.R., Rostkowski, M., Jensen, J.H.: PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7(2), 525–537 (2011). doi:10.1021/ct100578z
Gabb, H.A., Jackson, R.M., Sternberg, M.J.: Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 272(1), 106–120 (1997). doi:10.1006/jmbi.1997.1203
Kishore, R., Kaur, M.T.: Backpropagation algorithm: an artificial neural network approach for pattern recognition. Int. J. Sci. Eng. Res. 3(6), 1–4 (2012)
Pierce, B.G., Hourai, Y., Weng, Z.: Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLOS ONE 6(9), e24657 (2011). doi:10.1371/journal.pone.0024657
Acknowledgments
This research was supported by National Natural Science Fundation of China (31670725), and State Key Laboratory of Membrane Biology to Xinqi Gong. Experiments run on Renda Xing Cloud that currently has 64 physical nodes.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Yang, Y., Gong, X. (2017). Understanding Protein-Protein Interface Formation Mechanism in a New Probability Way at Amino Acid Level. In: Cai, Z., Daescu, O., Li, M. (eds) Bioinformatics Research and Applications. ISBRA 2017. Lecture Notes in Computer Science(), vol 10330. Springer, Cham. https://doi.org/10.1007/978-3-319-59575-7_36
Download citation
DOI: https://doi.org/10.1007/978-3-319-59575-7_36
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-59574-0
Online ISBN: 978-3-319-59575-7
eBook Packages: Computer ScienceComputer Science (R0)