Abstract
Scientists exploring a new area of research are interested to know the “hot” topics in that area in order to make informed choices. With exponential growth in scientific literature, identifying such trends manually is not easy. Topic modeling has emerged as an effective approach to analyze large volumes of text. While this approach has been applied on literature in other scientific areas, there has been no formal analysis of bioinformatics literature.
Here, we conduct keyword and topic model-based analysis on bioinformatics literature starting from 1998 to 2016. We identify top keywords and topics per year and explore temporal popularity trends of those keywords/areas. Network analysis was conducted to identify clusters of sub-areas/topics in bioinformatics. We found that “big-data”, “next generation sequencing”, and “cancer” all experienced exponential increase in popularity over the years. On the other hand, interest in drug discovery has plateaued after the early 2000s.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Scientific literature holds a rich record of the ever-changing landscape of thought and observations in a wide variety of domains. Within a particular domain, researchers are increasingly interested in exploring scientific literature to gain insights on how research develops and evolves over time [24]. For instance, this kind of analytical data-driven insight can benefit researchers as they delve into new areas by providing knowledge of current popular topics and how the focus on different topics has shifted through time [1, 24]. While the advent of digital publishing and open access science have led to greater access to scientific content, the sheer volume has made it very difficult for researchers to analyze literature at a high level and identify temporal trends in the evolution of research areas. [24]. This problem is particularly relevant in the thriving field of bioinformatics that encompasses several sub-areas garnering interest from biologists, computer scientists, and mathematicians.
Several approaches have been developed for analyzing text to identify semantic content, the most notable being topic modeling. Topic modeling is a text mining technique that identifies the hidden thematic/latent structure in collections of documents thereby allowing us to efficiently summarize large volumes of text [6]. Topic modeling algorithms take documents in a corpus and identify salient words grouping them to form ‘topics’. Each document in a corpus is represented as a probabilistic mixture of topics while each topic consists of a mixture of words. In this manner, topic modeling algorithms discover patterns in textual data via topic generation and use those topics to connect documents with similar content [1]. This approach of analyzing text has been used in disparate domains such as social sciences, business analytics, and computer science.
While there are several topic modeling algorithms [6, 10, 11], Latent Dirichlet Allocation (LDA) [6] is one of the most widely used approaches and has been shown to be effective at finding distinct topics from a corpus [7, 24]. In LDA, the topic distribution is assumed to have a Dirichlet prior unlike other algorithms such as LSA [10] and pLSA [11].
LDA model representation for W words over D documents with K topics [6]. The two boxes represent replicates with the outer box representing documents and the inner box representing topics and words within a document.
LDA is a generative statistical model that models each of D documents in a corpus as a mixture of K topics where each topic corresponds to a multinomial distribution of W words [6] (Fig. 1). Other parameters in the model are defined as follows:
-
\(\alpha \): Dirichlet prior on the topic distributions of each document
-
\(\beta \): Dirichlet prior on the word distributions of each word,
-
\(\theta _{d}\): Topic distribution for document d,
-
\(\varphi _{k}\): Word distribution for topic k,
-
\(z_{ij}\): Topic for the ith word in document j, and
-
\(w_{ij}\): A particular word.
One of the input parameters of the LDA algorithm is the number of topics (K) to be identified from the corpus. Several studies have developed approaches to determine the optimal number of topics [4, 22]. While there are likelihood based measures that help determine the right number of topics, these measures cannot be used alone to find the best model [3].
Here, we present our work on analyzing decades of Bioinformatics scientific literature to identify broad research themes and how those themes evolve across time. The goal of this work is to provide an exploration of different research areas within bioinformatics, identify “hot” areas and show how these areas interact with one another. We conduct a two-pronged analysis to achieve this goal. First, we analyze keywords and their popularity in each year to understand trends in popular research. A network of top keywords is built to identify clusters within these popular areas to observe interactions. Next, we apply topic modeling on abstracts to identify salient research themes at greater detail than keywords. These themes are complementary to themes identified from keywords. A network of topics is created to show how these research themes overlap and interact with each other. We explore temporal analysis of 10 curated topics to identify how research topics trend over time.
2 Related Work
Several studies have demonstrated the use of topic modeling to analyze scientific literature. Paul and Girju conducted analysis of literature in Computational Linguistics, and Education [17]. Their work shows how topics change over time in each field and how topics across fields are related. Similarly, Bolelli and Gilesb analyzed publications to identify research topics in computer science, influential authors, and trends related to those topics [7]. In a recent study, Kane et al. used topic models to compare the development of research on crops such as wheat, rice, sorghum, etc. [13]. Results from the topic models revealed interesting trends on how research on perennial crops was advancing and that is different from the progress on individual crops.
Much closer to our work is Altena et al.’s study on understanding the term big data from a text analysis of bio-medical literature [3]. While there are similarities in the literature corpus and techniques being applied, Altena et al.’s work differs from this study in that they restrict their study to big data literature in the bio-medical field while we analyze all areas of bioinformatics literature. In addition, we aim to search for over-arching patterns and trends in bioinformatics rather than focusing on one particular concept such as big data. Lastly, Suominen et al. performed topic modeling using LDA on scientific literature from Web of Science to compare how latent topics identified by LDA correlate with human assigned keyword categorization. The only use of topic models relating to bioinformatics to the best of our knowledge has been to answer specific research questions such as cluster analysis on medical, biological genotyping data [23] and toxicogenomics data analysis [15]. There is a notable lack of topic modeling based text analysis aimed at the wide corpus of bioinformatics literature to identify salient research topics and their evolution over time. Our work here aims to fill this gap.
3 Methods
3.1 Data Collection - Creating the Corpus
Scientific literature for this study was obtained by searching the Scopus database (https://www.scopus.com/) using the search term “bioinformatics”. Scopus adds relevant index terms selected from controlled vocabularies to all publications (https://www.elsevier.com/__data/assets/pdf_file/0007/69451/scopus_content_coverage_guide.pdf). In addition to author keywords and titles, these index terms are used for searching. A list of publications matching the search term were retrieved using the Scopus API. Next, Scopus was queried to retrieve additional data such as authors, keywords, abstract, year of publication, and other metadata corresponding to the publication. For each publication, a document was created by concatenating the corresponding title, keywords, and abstract. A corpus of scientific literature was created by putting together documents corresponding to each publication.
3.2 Keyword-Based Analysis
Publications were analyzed based on their keywords in the following ways:
-
1.
The number of publications per year was examined to identify any significant trends in research output across years.
-
2.
The number of unique keywords observed in each publication year was extracted to explore correlations with the distribution of publication output.
-
3.
A list of 25 keywords selected from the top keywords per year was curated and temporal analysis of their popularity across years was conducted. The popularity of a keyword computed using its occurrence frequency across documents per year was normalized to the [0, 1] range using min-max scaling. This analysis identified research areas experiencing upward spikes and rise in popularity and those experiencing decline.
-
4.
A network of the top 25 keywords per year was built to explore relationships, inter-connectivity, and to identify clusters among these keywords. The network arranges the set of keywords into clusters and identifies intra- and inter-cluster interactions. Keywords in the network are weighted based on the prominence of their association with different publications. The larger the proportion of publications a keyword is associated with, the larger the keyword appears on the network. The network was constructed using Gephi (https://gephi.org/), an open source tool for network building and analysis. Clusters/communities in the keyword network are detected and optimized using the Louvain method [9]. After initial clusters are formed, the modularity optimization component further optimizes the clusters.
3.3 Topic Model Based Analysis
Latent Dirichlet Allocation was applied on the literature corpus - a collection of documents, one corresponding to each publication. 6 topic models were created using K (number of topics) in the range of 25 to 150 at increments of 25. These topic models were evaluated through manual examination. For each model, 20 top words per topic were examined to assess scientific coherence of the words as a set, overlap in topic words across topics, and human understandability. The selected model was used for all subsequent analyses.
After model selection, publications were analyzed based on their topics in the following ways:
-
1.
The top 10 salient words relevant to 10 curated topics in the model were extracted and reported. This report provides a descriptive view of the topic model and verifies if topics identified by the model match natural human perception of the sub-areas of research within biology/bioinformatics.
-
2.
A topic similarity network of all topics was built to identify topic clusters and their interplay. This allows for the identification of exciting clusters of research areas within bioinformatics.
4 Results
Searching for the term “bioinformatics” on the Scopus database resulted in 85,106 publications between the years of 1998 and 2016. When grouped by year, we see an upward trend in the number of publications per year (Fig. 2) except for years 2012 and 2013. Surprisingly, there appears to be a noticeable drop in publications in those two years.
4.1 Keyword-Based Analysis
We found 100,754 unique keywords across the 85,106 publications spanning across 18 years with an average of about 3 keywords per publication. The trend in the distribution of unique keywords in publications per year (Fig. 2) is very similar to the distribution of yearly publication numbers.
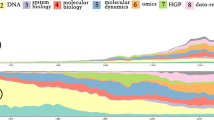
Temporal Keyword Trends. We manually curated 25 interesting keywords from top keywords in each year. Figure 3 shows the popularity trends of these 25 curated keywords. “big data”, “proteomics”,“rna seq”, “cancer”, “next generation sequencing”, and “transcriptomics” are among the areas that exhibit an increasing presence in publications over the last decade. It is interesting to see the emergence of big data applications within bioinformatics around 2010 accompanied by an exponential increase in relevant publications. “rna seq”, or rna sequencing, is another area that emerged during the later parts of the past decade and has emerged as a very popular research area. Unsurprisingly, the trend of “next generation sequencing” is similar to “rna seq”. Overall, “next generation sequencing techniques”, “cancer informatics”, “biomarkers”, “metabolomics”, “mirna”, “machine learning”, and “big data” are promising areas of research based on these trends. The emphasis on “cancer”, “biomarkers”, and “big data” indicate that health informatics is a sought after specialization. However, surprisingly, the same positive trend is not observed in the area of “drug discovery” which has plateaued over time. “functional genomics”, “ontologies”, and “neural networks” show mixed trends.
Keyword Network. The network built using the top 25 keywords per year comprises 6 clusters shown in blue, pink, purple, green, brown, and grey (Fig. 4). It is evident that the blue cluster is central to the network with substantial overlap with other clusters. For lack of space, we only show the central blue and the green cluster in greater detail (Figs. 5 and 6). The blue cluster (Fig. 5) is largely focused on health informatics - in particular the study of different types of cancer such as “colorectal”, “prostate”, “breast”, etc. The cluster accurately identifies that microarray and gene expression analyses have been significant contributors to the study of cancer in the past decades [8, 12, 18]. It also hints at more recent approaches to cancer analytics which include using “gene ontology”, “text mining”, and machine learning approaches such as “clustering”, etc. [14, 20].
The green cluster (Fig. 6) focuses largely on sequence analysis and alignment using algorithms and techniques from graph theory. The green cluster contains certain nodes that are a bit distant from the rest of the cluster. These words include “MPI”, “hadoop”, “mapreduce”, “cuda”, and “membrane”, “cloud computing”. Interestingly, all these words pertain to big-data approaches that have recently come into play to analyze high throughput data from next generation sequencing approaches [19, 21]. As sequencing data becomes more and more complex and voluminous, we can expect these words to become more central to this cluster over time.
The brown cluster focuses on computational techniques such as data mining, machine learning, feature selection for drug design and discovery, protein-structure prediction, pattern recognition, structural bioinformatics, etc. Moving on to the pink cluster, we see “data integration”, “database”, “semantic web”, and ontologies being used for the study of phenotypes, evolution, and phylogenies. This cluster points to the increasing applications of ontologies and data integration for the study of evolutionary phenotypes [16]. The grey cluster is largely related to proteomics, systems biology, functional genomics, analysis of microrna etc. The purple cluster is related to next generation sequencing, gene expression analyses, genomics, transcriptome, and genetics.
5 Topic Model-Based Analysis
Six topic models were created using number of topics (K) \(\in \) [25, 50, 75, 100, 125, 150]. After careful manual evaluation of the topics and their top words, the model built using 50 topics was selected based on topic coherence and human understanding. Increasing the number of topics would make each individual topic more specific and might increase overlap between topics. Decreasing the number of topics, would result in more high-level abstract topics. A snapshot of the 50 topic model is shown in Table 1 by illustrating salient words in 10 curated topics.
Topic Similarity Network. Next, we built a topic similarity network of all topics. In this network, nodes indicate topics represented by topic number and edges represent similarity between topics computed using the complement of Hellinger distance [5] between the probability distributions of two topics. The topic similarity network reveals four clusters (shown in blue, green, purple, and brown) (Fig. 7).
Topics in the purple cluster correspond mainly to health informatics, clinical informatics, specifically focusing on cancer informatics. The topics in this cluster are characterized by words such as “drug discovery”, “tumor”, “mirna studies”, “gene expression”, “association studies”, “target cells”, “differentially expressed”, “phage”, “genetic variants”, “biomarkers”, “early treatment”, “clinical diagnosis”, etc. The topics hint at ontologies, pathways, networks, text mining, and association studies as some of the computational tools used in this area of research.
Sequence alignment, sequence similarity, and other related applications are seen prominently in the green cluster. Other top areas in this cluster include phylogenetic trees, evolutionary algorithms, protein structure and prediction, protein interactions, and distributed computing. The blue cluster represents research in proteomics, genome sequencing, annotation, and assembly tools. Other areas represented in this cluster include metabolomics, protein structures, mass spectrometry, community software, and genome databases. Interestingly, the brown cluster which contains only two topics representing studies on water quality and treatment, is an outlier to the other clusters. Prominent words in these two topics include ph level, removal, water quality, nanoparticles, adsorption, iron, and concentration indicating work on water treatment advances using adsorption [2]. It is not surprising that these topics have little similarity with the other areas.
Overall, these topics indicate research areas such as health and cancer informatics, proteomics, genome annotation and assembly, sequence alignment, and the computational techniques used in each of these areas.
6 Conclusion
In this study, we conducted scientific literature analysis of bioinformatics publications from 1998 to 2016 using keyword and topic modeling based analysis. We discovered research areas within bioinformatics that are experiencing a rise in popularity and those witnessing waning interest. The trends show that there is increasing research in cancer informatics and that cancer research has shifted towards using big data techniques in recent years. The presence of big data techniques can also be seen in other areas such as sequence alignment and genome annotation. Machine learning, feature selection, network analysis, ontologies, data mining, distributed computing, parallel computing, hadoop, web applications, and community databases are some of the prominent computational techniques seen in bioinformatics.
References
Alghamdi, R., Alfalqi, K.: A survey of topic modeling in text mining. Int. J. Adv. Comput. Sci. Appl. (IJACSA) 6(1) (2015)
Ali, I., Gupta, V.: Advances in water treatment by adsorption technology. Nat. Protoc. 1(6), 2661–2667 (2006)
Altena, A.J., Moerland, P.D., Zwinderman, A.H., Olabarriaga, S.D.: Understanding big data themes from scientific biomedical literature through topic modeling. J. Big Data 3(1), 23 (2016)
Arun, R., Suresh, V., Veni Madhavan, C.E., Narasimha Murthy, M.N.: On finding the natural number of topics with latent dirichlet allocation: some observations. In: Zaki, M.J., Yu, J.X., Ravindran, B., Pudi, V. (eds.) PAKDD 2010. LNCS, vol. 6118, pp. 391–402. Springer, Heidelberg (2010). doi:10.1007/978-3-642-13657-3_43
Beran, R.: Minimum hellinger distance estimates for parametric models. Ann. Stat. 5, 445–463 (1977)
Blei, D.M., Ng, A.Y., Jordan, M.I.: Latent Dirichlet allocation. J. Mach. Learn. Res. 3(Jan), 993–1022 (2003)
Bolellia, L., Gilesb, S.: What is trendy? generative models for topic detection in scientific literature
Cheang, M.C., van de Rijn, M., Nielsen, T.O.: Gene expression profiling of breast cancer. Annu. Rev. Pathmechdis. Mech. Dis. 3, 67–97 (2008)
De Meo, P., Ferrara, E., Fiumara, G., Provetti, A.: Generalized louvain method for community detection in large networks. In: 2011 11th International Conference on Intelligent Systems Design and Applications (ISDA), pp. 88–93. IEEE (2011)
Deerwester, S., Dumais, S.T., Furnas, G.W., Landauer, T.K., Harshman, R.: Indexing by latent semantic analysis. J. Am. soc. Inf. Sci. 41(6), 391 (1990)
Hofmann, T.: Probabilistic latent semantic indexing. In: Proceedings of the 22nd annual international ACM SIGIR conference on Research and development in information retrieval, pp. 50–57. ACM (1999)
Hoopes, L.: Genetic diagnosis: DNA microarrays and cancer. Nat. Educ. 1(1), 3 (2008)
Kane, D.A., Rogé, P., Snapp, S.S.: A systematic review of perennial staple crops literature using topic modeling and bibliometric analysis. PloS One 11(5), e0155788 (2016)
Kourou, K., Exarchos, T.P., Exarchos, K.P., Karamouzis, M.V., Fotiadis, D.I.: Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 13, 8–17 (2015)
Lee, M., Liu, Z., Huang, R., Tong, W.: Application of dynamic topic models to toxicogenomics data. BMC Bioinf. 17(13), 368 (2016)
Manda, P., Balhoff, J.P., Lapp, H., Mabee, P., Vision, T.J.: Using the phenoscape knowledgebase to relate genetic perturbations to phenotypic evolution. Genesis 53(8), 561–571 (2015)
Paul, M.J., Girju, R.: Topic modeling of research fields: an interdisciplinary perspective. In: RANLP, pp. 337–342 (2009)
Perez-Diez, A., Morgun, A., Shulzhenko, N.: Microarrays for cancer diagnosis and classification. In: Mocellin, S. (ed.) Microarray Technology and Cancer Gene Profiling, pp. 74–85. Springer, New York (2007)
Vijayakumar, S., Bhargavi, A., Praseeda, U., Ahamed, S.A.: Optimizing sequence alignment in cloud using hadoop and MPP database. In: 2012 IEEE 5th International Conference on Cloud Computing (CLOUD), pp. 819–827. IEEE (2012)
Wu, T.J., Schriml, L.M., Chen, Q.R., Colbert, M., Crichton, D.J., Finney, R., Hu, Y., Kibbe, W.A., Kincaid, H., Meerzaman, D., et al.: Generating a focused view of disease ontology cancer terms for pan-cancer data integration and analysis. Database 2015, bav032 (2015)
Xue, Q., Xie, J., Shu, J., Zhang, H., Dai, D., Wu, X., Zhang, W.: A parallel algorithm for dna sequences alignment based on MPI. In: 2014 International Conference on Information Science, Electronics and Electrical Engineering (ISEEE), vol. 2, pp. 786–789. IEEE (2014)
Zhao, W., Chen, J.J., Perkins, R., Liu, Z., Ge, W., Ding, Y., Zou, W.: A heuristic approach to determine an appropriate number of topics in topic modeling. BMC Bioinf. 16(13), S8 (2015)
Zhao, W., Zou, W., Chen, J.J.: Topic modeling for cluster analysis of large biological and medical datasets. BMC Bioinf. 15(11), S11 (2014)
Zhou, H.K., Yu, H.M., Roland, H.: Topic discovery and evolution in scientific literature based on content and citations. Frontiers 1 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2017 The Author(s)
About this paper
Cite this paper
Hahn, A., Mohanty, S.D., Manda, P. (2017). What’s Hot and What’s Not? - Exploring Trends in Bioinformatics Literature Using Topic Modeling and Keyword Analysis. In: Cai, Z., Daescu, O., Li, M. (eds) Bioinformatics Research and Applications. ISBRA 2017. Lecture Notes in Computer Science(), vol 10330. Springer, Cham. https://doi.org/10.1007/978-3-319-59575-7_25
Download citation
DOI: https://doi.org/10.1007/978-3-319-59575-7_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-59574-0
Online ISBN: 978-3-319-59575-7
eBook Packages: Computer ScienceComputer Science (R0)