Abstract
Segmentation of the liver from abdominal CT images is a prerequisite for computer aided diagnosis. However, it is still a challenging task due to the low contrast of adjacent organs and the varying shapes between subjects. In this paper, we present a liver segmentation framework based on prior model using level set. We first weight all of the atlases in the training volumes by calculating the similarities between the atlases and the test image to generate a subject-specific probabilistic atlas. Based on the generated atlas, the most likely liver region (MLLR) of the test image is determined. Then, a rough segmentation is performed by a maximum a posteriori classification of probability map. The final result is obtained by applying a shape-intensity prior level set inside the MLLR with narrowband. We use 15 CT images as training samples, and 15 exams for evaluation. Experimental results show that our method can be good enough to replace the time-consuming and tedious manual approach.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Recent studies on liver segmentation can be classified into two types: The first type segments liver by use of pure image information, such as thresholding, clustering, and region growing. The deficiency of these schemes is the tendency to leak into adjacent organs because of their similarity in grey levels.
Another type is model-based methods. These methods aim to overcome the limit of the techniques mentioned above by combining local and global prior knowledge of liver shape. And they can be further divided into three categories: active and statistical shape models, atlas-based segmentation, and level set-based segmentation. In active shape model based approaches, statistical shape models (SSM) are usually employed to learn the shape-prior models [1, 2]. However, SSM approaches tend to overly constrain the shape deformations and overfit the training data, because of the small size of training samples. In atlas-based studies. Oda et al. [3] divided an atlas database into several clusters to generate multiple probabilistic atlases of organ location. In recent work, subject-specific probabilistic atlas (PA) based methods have been proposed [4], which is generated by registering multiple atlases to every new target image. In level set-based studies, Oliveira et al. [5] proposed a gradient-based level set model with a new optimization of parameter weighting for liver segmentation. Li et al. [6] suggested a combination of gradient and region properties to improve level set segmentation.
In this paper, an automatic scheme on segmenting liver from abdominal CT images are proposed. Firstly, the similarities between all atlases and the test volume are calculated; Secondly, the most likely liver region (MLLR) of the test image is constructed, and a maximum a posteriori (MAP) classification of probability map is depicted; Then, a shape-intensity prior level set is applied to produce the final liver segmentation inside the MLLR. Finally, for constructing shape-intensity models of the liver, 15 CT samples are used as training samples, and 15 test volumes are used for evaluating the proposed scheme.
2 Description of the Method
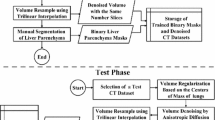
Our research consists of a training phase and a testing phase. We will explain how to segment the liver automatically from abdominal CT images stage by stage, with shape-intensity prior models using level set technology. Figure 1 depicts the flowchart of the segmentation scheme.
2.1 Image Preprocessing
In the image preprocessing stage. Firstly, All the volumes are regularized to reduce the changes between individuals according to the centers of the lungs. And then, the traditional binary thresholding algorithm is used for the lung parenchyma extraction. Thirdly, for reducing noise and preserving the liver contour, an anisotropic diffusion filter is applied, followed by an isotropic resampling process based on trilinear interpolation technology. In this way, z-axis is resampled to the same number of slices, since a fixed number of the input vectors are required by principal component analysis (PCA) algorithm.
2.2 Liver Rough Segmentation
Liver rough segmentation can be divided into three stages: (1) Most likely liver region construction; (2) Probability map based classification; (3) Non-liver elimination.
Most Likely Liver Region Construction. For constructing the subject-specific weighted probabilistic atlas, the image similarities between all chosen atlases and the test image are calculated, and then, arranging them in descending order, which would be evaluated by normalized cross correlation (NCC) [7]:
where A and T denote the atlas and the test image, respectively, and \(\bar{A}\), \(\bar{T}\) represent the average intensity of A and T. i, j and k denote the coordinate value of x, y , z axes, and each atlas is assigned with a weight as
where n represents the index of atlas, w(n) is the weight of the n-th atlas, order(n) denotes the order of the n-th atlas, and N is the total number of atlases. It is evident that, the more similar to the target image, the bigger order and weight the atlases would be assigned. Then we defined the weighted probabilistic atlas \(W_p(lv)\) of liver at voxel p as:
and \(s(x,x')\) represents the similarity function, if \(I=I'\), then, \(s(I,I')=1\); otherwise, \(s(I,I')=0\). i is the atlas index, and \(L_p^i\) denotes the voxel p on atlas i. In this way, the subject-specific weighted PA is constructed, and then, we obtain the most likely liver region (MLLR) by appropriate thresholding processing.
Probability Map Based Clustering. After the construction of MLLR, we divide the CT intensities inside the MLLR into six categories: liver, heart, right kidney, spleen, bone and background, and we utilized the intensity histograms of the six categories in the training stage. The likelihood for each category is defined by p(I(x)|q) (q represents the six organs.), which can be calculated by convolving their histograms. Then according to the Beyesian theorem, we get the following equation:
where P(lv) denotes the liver probability map, and I(x) represents the CT intensity of position x. By a MAP classification of probability map, we get a rough segmentation of the liver.
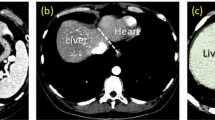
Non-liver Region Elimination. The rough segmentation of liver could results in some segmentation errors, due to the similar intensities with its adjacent organs. Based on the fact that liver is the largest organ in the MLLR, thus we applied a morphological operation to fill the holes, followed by the connected component analysis to eliminate the non-liver region. A rough segmentation process is shown in Fig. 2.
2.3 Refinement of Rough Segmentation
In this section, we would refine the rough segmentation result through four steps: (1) Maximum a posteriori (MAP) framework construction; (2) Construction of prior model; (3) Formulation of level set; (4) Evolution of the liver surface.
Maximum a Posteriori (MAP) Framework Construction. For making the best use of image information, such as the shape and the grey level, we derived the MAP framework referring to the conclusion proposed by Yang et al. [8]:
where I denotes the target image with object \(\varphi \), and \(I_{\varphi }\) is the synthetic model of image I; \(\hat{I}_{\varphi }\) is the segmentation of \(I_{\varphi }\). In this case, we assume the synthetic image \(I_{\varphi }\) is quite close to the real image I ( \( I \approx I_{\varphi }\) ), and \(p(I|\varphi ,I_{\varphi })\) is the term based on the image intensity information. In three-dimensional image, assuming intensity homogeneity within the object, the following imaging model is derived [9]:
here, function \(p(\varphi , I)\) combined shape \(\varphi \) with intensity I, and \(a_1\), \(\sigma _1\), \(a_2\) , \(\sigma _2\) denote the average gray level and the variance, inside and outside \(\varphi \), respectively.
Construction of Prior Model. For constructing the shape-intensity based prior model with the training samples, the technology proposed by Cootes et al. [10] is utilized, Therefore, an estimate of the shape-intensity pair \([\bar{\varPsi }^T,\bar{I}^T]^T\) can be represented by k principal components and a k dimensional vector of coefficients \(\alpha \):
where \(\varPsi \) is the level set function, I represents the sample images, and \(U_k\) represents the matrix consisting of the first k columns of matrix U. Figure 3 demonstrates the variation of shape-intensity on the first three modes.
Under the assumption of a Gaussian distribution of a shape-intensity pair represented by \(\alpha \), the joint probability of a certain shape \(\varphi \) and the related image intensity I, \(p(\varphi ,I)\) can be represented by
Similar to [9], a boundary smoothness regularization term is incorporated to add robustness against noise data: \(p_B(\varphi )=e^{-\mu \oint _{\varphi } {\text{ d }s}}\), where \(\mu \) is a scalar factor. By adding the above regularizing term, the shape-intensity probability \(p(\varphi ,I)\) can be approximated by a product of the following probabilities:
Substitute Eqs. 6 and 9 into Eq. 5, we derive \(\hat{\varphi }\) by obtaining the minimizer of the following energy functional \(E(\varphi )\) shown below
In this paper, the minimization problem of energy functional \(E(\varphi )\) would be formulated and solved by the level set strategy.
Formulation of Level Set. For formulating the level set prior model, we replace \(\varphi \) with \(\phi \) in the energy functional of Eq. 10, and for calculating the associated Euler-Lagrange equation, we minimize E with reference to \(\phi \). According to the gradient descent method with artificial time \(t\geqslant 0\), the evolution equation in \(\phi (t,{x})\) is obtained (Hamilton-Jacobi equation):
where G(.) is used for representing matrix in column scanning, while g(.) denotes its reverse operation, and \(U_{k1}\) and \(U_{k2}\) represent the upper and lower half of the matrix \(U_k\), respectively.
Evolution of the Liver Surface. The evolving of the surface inside MLLR can be summarized by the following six steps:
step 1: Initialize the curved surface of the liver \(\phi \) at time t;
step 2: Calculate \(p(\alpha )\) using PCA;
step 3: Calculate \(a_1(\phi ^t), \sigma _1(\phi ^t)\) and \(a_2(\phi ^t),\sigma _2(\phi ^t)\);
step 4: Update \(\phi ^{t+1}\);
step 5: Repeated from step 1;
step 6: If convergence, algorithm stop, otherwise, continue from step 3.
3 Experimental Results
In this section, we perform quantitative evaluations and demonstrate segmentation results via testing on 15 volumes provided by our clinic partner. Performance results are compared with Linguraru’s method [11]. 15 training samples with reference from Weihai Municipal Hospital are used to construct the shape-intensity models of the liver, while the accuracy of scheme was quantitatively performed using the following three measures: average symmetric surface distance (ASD), root mean square symmetric surface distance (RMSD), and Jaccard similarity coefficient (JSC).
Figure 4 shows some typical segmentation results between Linguraru’s method and ours, and our method obtain a better performance in some difficult cases.
Figure 5 shows the ASD error between Linguraru’s method and ours. The average ASD error is 1.56 \(\pm \) 0.13 mm (min 1.21, max 2.47) by Linguraru’s method, and 1.18 \(\pm \) 0.12 mm (min 0.82, max 2.12) by ours.
Figure 6 shows the RMSD errors between Linguraru’s method and ours. The average RMSD error is 2.40 \(\pm \) 0.20 mm (min 1.91, max 3.41) by Linguraru‘s method, and 1.87 \(\pm \) 0.23 mm (min 1.42, max 3.01) by ours.
Table 1 shows the Jaccard similarity coefficient (JSC) on the 15 exams among Lingurarus method, ours, and a manual segmentation from a senior radiologist. All the comparison results above indicate that, our segmentation scheme achieved a better segmentation accuracy than Lingurarus method, and is closer to the level of radiologist.
4 Conclusion
In this paper, a shape-intensity prior level set method is applied for segmenting liver from contrast-enhanced CT images, combined with probabilistic atlas and probability map constrains. We used 15 training samples of abdomen CT to construct liver shape-intensity prior models, and compare our approach with Linguraru’s method on 15 testing volumes. Results show that our method is a good promising tool on liver segmentation.
References
So, R., Chung, A.: Multi-level non-rigid image registration using graph-cuts. In: ICASSP 2009, Taipei, pp. 397–400 (2009)
Wimmer, A., Soza, G., Hornegger, J.: A generic probabilistic active shape model for organ segmentation. In: Yang, G.-Z., Hawkes, D., Rueckert, D., Noble, A., Taylor, C. (eds.) MICCAI 2009, Part II. LNCS, vol. 5762, pp. 26–33. Springer, Heidelberg (2009)
Okada, T., Yokota, K., Hori, M., Nakamoto, M., Nakamura, H., Sato, Y.: Construction of hierarchical multi-organ statistical atlases and their application to multi-organ segmentation from CT images. In: Metaxas, D., Axel, L., Fichtinger, G., Székely, G. (eds.) MICCAI 2008, Part I. LNCS, vol. 5241, pp. 502–509. Springer, Heidelberg (2008)
Wolz, R., Chu, C., Misawa, K., Mori, K., Rueckert, D.: Multi-organ abdominal CT segmentation using hierarchically weighted subject-specific atlases. In: Ayache, N., Delingette, H., Golland, P., Mori, K. (eds.) MICCAI 2012, Part I. LNCS, vol. 7510, pp. 10–17. Springer, Heidelberg (2012)
Oliveira, D.A., Feitosa, R.Q., Correia, M.M.: Segmentation of liver, its vessels and lesions from CT images for surgical planning. Biomed. Eng. Online 10, 30 (2011)
Li, B.N., Chui, C.K., Chang, S., Ong, S.H.: Integrating spatial fuzzy clustering with level set methods for automated medical image segmentation. Comput. Biol. Med. 41, 1–10 (2011)
Oda, M., Nakaoka, T., Kitasaka, T., Furukawa, K., Misawa, K., Fujiwara, M., Mori, K.: Organ segmentation from 3D abdominal CT images based on atlas selection and graph cut. In: Yoshida, H., Sakas, G., Linguraru, M.G. (eds.) Abdominal Imaging. LNCS, vol. 7029, pp. 181–188. Springer, Heidelberg (2012)
Yang, J., Duncan, J.S.: 3D image segmentation of deformable objects with joint shape-intensity prior models using level sets. Med. Image Anal. 8, 285–294 (2004)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Image Process. 10, 266–277 (2001)
Cootes, T.F., Hill, A., Taylor, C.J., Haslam, J.: The use of active shape models for locating structures in medical images. In: Barrett, H.H., Gmitro, A.F. (eds.) IPMI 1993. LNCS, vol. 687, pp. 33–47. Springer, Heidelberg (1993)
Linguraru, M.G., Sandberg, J.K., Li, Z., Pura, J.A., Summers, R.M.: Atlas-based automated segmentation of spleen and liver using adaptive enhancement estimation. In: Yang, G.-Z., Hawkes, D., Rueckert, D., Noble, A., Taylor, C. (eds.) MICCAI 2009, Part II. LNCS, vol. 5762, pp. 1001–1008. Springer, Heidelberg (2009)
Acknowledgments
This work was supported by Scientific Research Fund of Heilongjiang Provincial Education Department (No. 12541164).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Wang, J., Cheng, Y. (2015). Automatic Liver Segmentation Scheme Based on Shape-Intensity Prior Models Using Level Set . In: Zhang, YJ. (eds) Image and Graphics. ICIG 2015. Lecture Notes in Computer Science(), vol 9217. Springer, Cham. https://doi.org/10.1007/978-3-319-21978-3_55
Download citation
DOI: https://doi.org/10.1007/978-3-319-21978-3_55
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-21977-6
Online ISBN: 978-3-319-21978-3
eBook Packages: Computer ScienceComputer Science (R0)