Abstract
Multithreaded wavefront-based alignment procedure is used in the PSS-SQL language that allows for flexible scanning of databases of protein secondary structures and finding similarities among protein molecules. Efficiency of the process depends on several factors, including the way how the similarity matrix, calculated during the process, is divided into areas, the number of CPU cores possessed by the computer hosting the database with PSS-SQL extension, and structural patterns submitted by users in PSS-SQL queries. In this paper, we show how we achieved consensus values of area sizes for the multithreaded wavefront-based alignment procedure by a series of experimental trials. Availability: PSS-SQL extension for Microsoft SQL Server database management system can be downloaded from PSS-SQL project home page available at: http://www.zti.aei.polsl.pl/w3/dmrozek/science/pss-sql.htm.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Proteins
- Secondary structure
- Structural patterns
- Structural bioinformatics
- Similarity searching
- Structural alignment
- Databases
- SQL
- Relational databases
- Query language
1 Introduction
Proteins, molecules of life, have usually complex construction that is described on four representation levels, from primary to quaternary structure [5]. Protein secondary structure as a representation level describes protein construction in terms of regularly occurring shapes, including α-helices, β-strands, loop, turns, or coils (Fig. 1a), that protein amino acid chain can adopt in some of its regions. This representation level is very important for studying potential functions of protein molecules in living cells, determination of domain boundaries, topology recognition, and family assignment. These processes are usually realized through scanning databases of protein structures and finding similarities between the specified structural pattern and candidate database structures. We have developed a dedicated query language called PSS-SQL, which allows to perform such similarity searches, when protein secondary structures are collected in relational database [3].
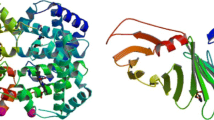
Crystal structure of the CAS1-CAS2 complex from Escherichia Coli [12], PDB ID: 4P6I, chain A: (a) tertiary (3D) structure with secondary structures (spiral α-helices, wavy β-strands, and loops) exposed in RasMol [14], (b) two representations of secondary structures in relational databases used by PSS-SQL: (top) full, showing types of secondary structures for corresponding residues, and (bottom) compressed, describing types and lengths of secondary structures identified; types of secondary structures: H – α-helix, E – β-strand, C – loop, turn or coil, U – undetermined structure.
1.1 PSS-SQL for Scanning Protein Secondary Structures in RDBMS
PSS-SQL (Protein Secondary Structure - Structured Query Language) [10, 11] is a query language that allows execution of similarity searches against sequences of protein secondary structures stored in relational database management systems (RDBMS). PSS-SQL was initially developed in Institute of Informatics at the Silesian University of Technology in Gliwice, Poland in 2009 and later enriched by several improvements. Technically, PSS-SQL is an extension to the Transact-SQL language implemented in Microsoft SQL Server RDBMS. The great power of PSS-SQL lies in the fact that it is a declarative language. While writing PSS-SQL queries, programmers and bioinformaticians, equipped with a library of functions and procedures extending standard searching capabilities of the database engine, specify the structural pattern of a protein for which they want to find similar proteins, provide what they want to display, where the data are stored, i.e. in which tables of the database, and how to filter them. For example, a simple SELECT statement, which is used to retrieve and display proteins that are similar to the given structural pattern, may have the following form:

This sample query returns Accession Numbers (AC) and names of proteins from Staphylococcus aureus having the length greater than 150 residues and structural region containing β-strand of the length from 1–10 elements, optional loop up to 5 elements, an α-helix of the length 5 to 6 elements, optional loop up to 5 elements, a β-strand of the length 1 to 10 elements and a 5 element loop. The structural region is defined by structural pattern e(1;10),c(0;5),h(5;6),c(0;5),e(1;10),c(5) passed to the sequenceMatch table function invoked in the FROM clause (dbo is a database schema name). Secondary structures are stored in the secondary column of the ProteinTbl in the form presented in Fig. 1b (top). Two additional columns, namely matchingSeq and already mentioned secondary, which are listed in the SELECT clause, show the exact pattern sequence that was matched to a candidate protein from the database and full secondary structure of the candidate protein. The matchingSeq column is returned by the sequenceMatch function.
1.2 Multithreaded Wavefront-Based Alignment
Due to the approximate nature of the similarity searching appropriate dynamic programming alignment procedure underlies all similarity searches and matches realized in PSS-SQL queries. More efficient scanning is achieved when the alignment procedure is preceded by multiple scanning of the segment index (MSSI) created for the table column storing secondary structures [10]. The index uses the representation of protein secondary structures shown in Fig. 1b (bottom). The alignment procedure makes use of results of the multiple scanning of the segment index. Therefore, this variant of the alignment procedure is marked as +MSSI.
In the +MSSI variant of the alignment the candidate database sequence of secondary structure S C is represented as follows:
where single element \( s_{j}^{C} \in \{ H,E,C,U\} \) corresponds to single secondary structure (segment) identified in the protein, \( l_{j} \) is the length of the j th segment measured in residues, and n is the number of segments. Example of such a sequence is shown in Fig. 1b (bottom).
Query protein structure S Q is represented as follows:
where single element \( s_{i}^{Q} \in \{ H,E,C,U\} \) corresponds to single secondary structure (segment) of the structural pattern provided by a user, \( l_{i} ,u_{i} \) are lower and upper limits for the length of the i th segment, both measured in residues, and m is the number of segments in the query pattern. Example of such defined pattern is shown in the sample PSS-SQL query presented in Sect. 1.1.
In the +MSSI variant of the alignment procedure, we calculate the similarity matrix D according to the following formulas.
where d i,j is the matching degree between elements of both sequences:
where \( \omega_{ + } \) is a matching award, and \( \omega_{ - } \) is a mismatch penalty. Additional matrices E and F are used for fast calculation of horizontal and vertical gaps:
for:
where: \( \sigma \) is the penalty for opening a gap in the alignment, and \( \delta \) is the penalty for extending the gap.
Since the alignment procedure is computationally costly, we have developed the multithreaded (+MT) implementation of the procedure. This implementation improves efficiency of the computational procedure, while the computational complexity of the alignment algorithm remains unchanged.
While performing the multithreaded, pairwise alignments the search engine of the PSS-SQL language calculates the similarity matrix D. Particular cells of the matrix depend on each other (each cell D i,j can be calculated only, if cells D i-1,j-1 , D i, j-1 , D i-1,j have already been calculated), so calculations must occur in a particular order. To avoid costly synchronizations of threads, the similarity matrix is divided into areas, which are assigned to working threads. Calculations are performed diagonally for areas, according to the wavefront approach [1, 6], as it is presented in Fig. 2.
Wavefront-based calculation of similarity matrix divided into areas (A u,v ). Compressed sequences of secondary structures (described by type and length of the structure) are located along vertical and horizontal edges of the matrix. We assumed that query protein structural pattern is defined precisely. Arrows show dependences between calculated areas. Areas already calculated are marked in grey, the area currently being calculated is marked in orange (A 2,2 ). Steps of the wavefront approach are marked by dashed lines.
While details of the alignment algorithm and general conclusions on performance of PSS-SQL queries were reported in [7, 10], the question remains how the size of the area influences the efficiency of PSS-SQL queries. Moreover, there are additional three factors that influence efficiency and should be taken into considerations: (1) the number of CPU cores possessed by the computer hosting the DBMS with the PSS-SQL extension, (2) structural patterns submitted by users in PSS-SQL queries, and (3) characteristics of the data stored in the database. Since each user may work on its own repository and we are unable to predict the characteristics of data collected in it, we decided to investigate the first and the second of the mentioned factors. In this paper, we show how we achieved consensus values of area sizes for both variants of the multithreaded wavefront-based alignment procedure by a series of experimental trials.
2 Consensus on the Area Size
Area sizes for the +MT+MSSI variant of the alignment procedure were chosen experimentally taking into account various patterns and varying number of CPU cores. Tests were performed on the Microsoft SQL Server 2012 EE working on nodes of the virtualized cluster controlled by the HyperV hypervisor hosted on Microsoft Windows 2008 R2 Datacenter Edition 64-bit. The host server had the following parameters: 2x Intel Xeon CPU E5620 2.40 GHz, RAM 92 GB, 3x HDD 1 TB 7200 RPM. Cluster nodes were configured to use from 1 up to 4 CPU cores and 4 GB RAM per node, and worked under the Microsoft Windows 2008 R2 Enterprise Edition 64-bit operating system. Tests were performed on the database storing 6 360 protein structures and for PSS-SQL queries containing patterns representing various classes:
-
class 1: short patterns and patterns frequently occurring in the database, e.g., c(10;20), h(2;5), c(2;40);
-
class 2: patterns with precisely defined regions, e.g., region e(15) in the sample pattern e(4;20), c(3;10), e(4;20), c(3;10), e(15), c(3;10), e(1;10), c(3;10), e(5;12);
-
class 3: patterns with unique regions, e.g., region h(243) in the sample pattern h(10;20), c(1;10), h(243), c(1;10), h(5;10), c(1;10), h(10;15);
-
class 4: patterns with undefined type of secondary structure (wildcard symbol ?) and with unlimited length of one of its regions (wildcard symbol *), e.g., region ?(1;30) and e(5;*), in the sample pattern c(10;20), h(2;5), c(2;40), ?(1;30), e(5;*).
Query patterns chosen for our tests had different characteristics and were representatives of possible patterns that can be entered by users. During experiments we have not observed any dependency between types of the secondary structures specified in patters and the execution time. The aim of the series of tests was to determine the possible best size of the area, which was assigned to every single thread. We have checked different area sizes for the following heights and widths: 1, 2, 3, …, 9. We tested popular configurations of 1, 2, 3, and 4 threads working in parallel, which corresponded to 1, 2, 3, and 4 CPU cores available for the database management system. This gave (9^2)∙4 = 324 combinations that were examined for each query pattern.
In Table 1 we can see execution times for the sample PSS-SQL query containing sample pattern from class 2. The class represents complex patterns that consist of many segments. As we can notice in Table 1 smaller area sizes (especially the size 1 × 1) result in higher execution times. Increasing the area size above 2 × 2 reduces the execution time. However, changes of the area size (above 3 × 3) do not affect the execution time significantly. The same tendency was observed in all tested cases, i.e. while testing various numbers of threads for different query patterns.
Table 2 shows relative execution times for the same pattern class. The table is presented as a heat map, where particular cells are colored using the red color (worst result), through yellow, and green color (best results). Results are expressed as a percentage - 0 % denotes the longest execution time, 100 % denotes the shortest execution time. Values are calculated according to the following expression:
where: t i,j is measured execution time of the sample query taken from Table 1 for the corresponding area size, max(t), min(t) are maximal and minimal execution times of the query (in Table 1).
The heat map (Table 2) reveals preferred and recommended area sizes (green) and those that should be avoided (red and orange). We have to remember that various query patterns and the number of possessed CPU cores may move the best point in any direction. Therefore, we have made these types of statistics, as presented in Tables 1 and 2, for all tested patterns in all tested classes for all tested n-core CPU configurations and area sizes. Then, in order to determine a universal area size for common patterns we have calculated the value of weighted arithmetic mean taking into account the participation of popular n-core processors in the market (n = 1, 2, 3, 4) and possible assignment of logical CPU cores in virtualized environments. We have arbitrary chosen the following values of weights: 15 % for 1-core CPUs (1 core = 1 thread execution), 40 % for 2-core CPUs, 5 % for 3-core CPUs, and 40 % for 4-core CPUs. Results are presented in Table 3. Best execution times (relative to the worst case) were obtained for the area width greater than 6. Height of the area has no significant impact, since compressed query sequences of secondary structures are usually short, while database sequences are relatively longer. In Fig. 3 we can see the histogram of the number of secondary structures identified in proteins stored in tested database. It shows that most of the proteins have less than 100 secondary structures (segments). Therefore, widths of whole similarity matrices after compression of sequences of secondary structures should be lower than 100 elements in most cases. Heights of whole similarity matrices depend on the query pattern. For example, the sample query pattern from pattern class 2 contains 9 segments, and consequently, the height of the similarity matrix is 9. Therefore, on the basis of our experiments, we have chosen 3 × 7 (H × W) for the area size, i.e., 3 for the query pattern and 7 for database sequence of secondary structures.
3 Related Works
Advantages of a declarative processing of biological data with the use of the SQL query language were noticed in the last decade, which resulted in the development of various SQL extensions. One of the first extensions was ODM BLAST [15], which was developed for Oracle RDBMS. ODM BLAST allows to align and match amino acid sequences of proteins (primary structures) and nucleotide sequences of DNA and RNA acids. Similarly, BioSQL [2], which makes use of particular modules of BioJava [13], provides a generic relational model for persistent storage of bio-molecular sequences, features, sequence and feature annotation, a reference taxonomy, and ontologies (or controlled vocabularies). Various bio-oriented projects may utilize the information by means of object-relational mapping (ORM). Recently reported P3D-SQL [9] extends Oracle PL/SQL capabilities by providing a set of functions and procedures that allow to perform structural alignments and superposition of 3D (tertiary) protein structures. Declarative processing of protein secondary structures is available by means of PSS-SQL [10, 11] and the query languages developed by Hammel and Patel [4] and Tata et al. [16]. These languages allow searching on the secondary structure of protein sequences. All mentioned projects confirm that for bio-database developers, highly skilled users, also those working in the domain of structural bioinformatics, the SQL language became an important communication interface.
4 Discussion and Concluding Remarks
By providing the PSS-SQL libraries to Microsoft SQL Server (version 2012 or higher) we have extended standard capabilities of the Transact-SQL language towards processing biological data. PSS-SQL extensions allow to store, index, process, compare, align and match protein structures based on their secondary structures in the relational database management system. These operations can be especially beneficial for database developers, data analysts, data scientists and programmers working in the domain of structural bioinformatics.
Efficiency of these operations, especially PSS-SQL queries, depend on several factors, including the internal configuration of the multithreaded wavefront-based alignment procedure, which is one of the two main phases of the query execution process. In this paper, we tried to show how we achieved the consensus area size for the alignment procedure by a series of experimental trials. We took into account two important factors that may influence the execution time, i.e., various query patterns (their lengths, uniqueness, vagueness) and the number of available CPU cores. These investigations allowed to develop efficient, domain-specific query language. Results of our experiments made PSS-SQL competitive to similar solutions implemented by Hammel and Patel [4] and Tata et al. [16], although, both mentioned solutions do not utilize alignment at all. Detailed performance evaluation and comparisons between mentioned query languages for protein secondary structures reported in Mrozek et al. [10] show that even with the computationally costly alignment the PSS-SQL is able to find protein similarities efficiently becoming a successful example of a DBMS-side processing. In such a way, PSS-SQL joins a narrow group of bio-oriented SQL extensions, such as BioSQL, ODM BLAST, and P3D-SQL, complementing the group with the important capability of finding similarities among proteins on the basis of their secondary structures. Thereby, PSS-SQL becomes a declarative, domain-specific query language for protein similarity searching and protein function identification on commonly available workstations without specialized equipment.
There is still a possibility to improve the efficiency of the PSS-SQL, e.g., by a parallelization of the alignment procedure on GPU devices. We have successfully developed such an alignment procedure for secondary structures in the GPU-CASSERT method [7, 8]. In the GPU-CASSERT the alignment of secondary structures alone (phase 1) takes just a fraction of seconds. However, the data must be prepared appropriately before the alignment begins, and data transfers between the host workstation and global memory of the GPU device also take some time.
Availability. PSS-SQL is free for scientific and testing purposes. It is available from PSS-SQL project home page at: http://zti.polsl.pl/w3/dmrozek/science/pss-sql.htm.
References
Anvik, J., MacDonald, S., Szafron, D., Schaeffer, J., Bromling, S., Tan, K.: Generating parallel programs from the wavefront design pattern. In: Proceedings of the 7th International Workshop on High-Level Parallel Programming Models and Supportive Environments (HIPS 2002), Fort Lauderdale, Florida, April 2002, pp. 1–8 (2002)
BioSQL. http://biosql.org/. Accessed 13 January 2015
Date, C.: An Introduction to Database Systems, 8th edn. Addison-Wesley, New York (2003)
Hammel, L., Patel, J.M.: Searching on the secondary structure of protein sequences. In: 28th International Conference on Very Large Data Bases VLDB 2002, Hong Kong, China, pp. 634–645 (2002)
Lesk, A.M.: Introduction to Protein Science: Architecture, Function, and Genomics, 2nd edn. Oxford University Press, New York (2010)
Liu, W., Schmidt, B.: Parallel design pattern for computational biology and scientific computting applications. In: Proceedings of the 2003 IEEE International Conference on Cluster Computing, Hong Kong, China, pp. 456–459 (2003)
Mrozek, D.: High-Performance Computational Solutions in Protein Bioinformatics. Springer Briefs in Computer Science. Springer International Publishing, Heidelberg (2014). http://dx.doi.org/10.1007/978-3-319-06971-5
Mrozek, D., Brożek, M., Małysiak-Mrozek, B.: Parallel implementation of 3D protein structure similarity searches using a GPU and the CUDA. J. Mol. Model. 20(2), 2067 (2014)
Mrozek, D., Małysiak-Mrozek, B., Adamek, R.: P3D-SQL: extending oracle PL/SQL capabilities towards 3d protein structure similarity searching. In: Ortuño, F., Rojas, I. (eds.) IWBBIO 2015, Part I. LNCS, vol. 9043, pp. 548–556. Springer, Heidelberg (2015)
Mrozek, D., Socha, B., Kozielski, S., Małysiak-Mrozek, B.: An efficient and flexible scanning of databases of protein secondary structures with the segment index and multithreaded alignment. J. Intell. Inf. Syst. (in press). http://dx.doi.org/10.1007/s10844-014-0353-0
Mrozek, D., Wieczorek, D., Małysiak-Mrozek, B., Kozielski, S.: PSS-SQL: protein secondary structure - structured query language. In: 32th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS 2010, Buenos Aires, Argentina, pp. 1073–1076 (2010)
Nunez, J.K., Kranzusch, P.J., Noeske, J., Wright, A.V., Davies, C.W., Doudna, J.A.: Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 21, 528–534 (2014)
Prlić, A., et al.: BioJava: an open-source framework for bioinformatics in 2012. Bioinformatics 28, 2693–2695 (2012)
Sayle, R.: RasMol, Molecular Graphics Visualization Tool. Biomolecular Structures Group, Glaxo Welcome Research & Development, Stevenage, Hartfordshire, 5 February 2013, 1998. http://www.umass.edu/microbio/rasmol/
Stephens, S.M., Chen, J.Y., Davidson, M.G., Thomas, S., Trute, B.M.: Oracle database 10 g: a platform for BLAST search and regular expression pattern matching in life sciences. Nucl. Acids Res. 33(Suppl 1), D675–D679 (2005). doi:10.1093/nar/gki114
Tata, S., Friedman, J.S., Swaroop, A.: Declarative querying for biological sequences. In: 22nd International Conference on Data Engineering, pp. 87–98. IEEE Computer Society, Atlanta (2006)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Mrozek, D., Małysiak-Mrozek, B., Socha, B., Kozielski, S. (2015). Selection of a Consensus Area Size for Multithreaded Wavefront-Based Alignment Procedure for Compressed Sequences of Protein Secondary Structures. In: Kryszkiewicz, M., Bandyopadhyay, S., Rybinski, H., Pal, S. (eds) Pattern Recognition and Machine Intelligence. PReMI 2015. Lecture Notes in Computer Science(), vol 9124. Springer, Cham. https://doi.org/10.1007/978-3-319-19941-2_45
Download citation
DOI: https://doi.org/10.1007/978-3-319-19941-2_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-19940-5
Online ISBN: 978-3-319-19941-2
eBook Packages: Computer ScienceComputer Science (R0)