Abstract
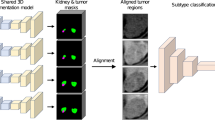
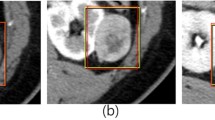
Deep learning (DL) has proven itself as a powerful tool to capture patterns that human eyes may not be able to perceive when looking at high-dimensional data such as radiological data (volumetric data). For example, the classification or grading of kidney tumors in computed tomography (CT) volumes based on distinguishable patterns is a challenging task. Kidney tumor classification or grading is clinically useful information for patient management and better informing treatment decisions. In this paper, we propose a novel DL-based framework to automate the classification of kidney tumors based on the International Society of Urological Pathology (ISUP) renal tumor grading system in CT volumes. The framework comprises several pre-processing techniques and a three-dimensional (3D) DL-based classifier model. The classifier model is forced to pay particular attention to the tumor regions in the CT volumes so that it can better interpret the surface patterns of the tumor regions to attain performance improvement. The proposed framework achieves the following results on a public dataset of CT volumes of kidney cancer: sensitivity 85%, precision 84%. Code used in this publication is freely available at: https://github.com/Balasingham-AI-Group/Classification-Kidney-Tumor-ISUP-Grade.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
- 2.
- 3.
- 4.
Area under the curve is a performance measurement for the classification problems. It tells how much the model is capable of distinguishing between classes.
References
Sung, H., et al.: Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 71(3), 209–249 (2021)
Molina, A.M., et al.: A phase 1b clinical trial of the multi-targeted tyrosine kinase inhibitor lenvatinib (e7080) in combination with everolimus for treatment of metastatic renal cell carcinoma (RCC). Cancer Chemother. Pharmacol. 73(1), 181–189 (2014)
Motzer, R.J., et al.: Dovitinib versus sorafenib for third-line targeted treatment of patients with metastatic renal cell carcinoma: an open-label, randomised phase 3 trial. Lancet Oncol. 15(3), 286–296 (2014)
Samaratunga, H., Gianduzzo, T., Delahunt, B.: The ISUP system of staging, grading and classification of renal cell neoplasia. J. Kidney Cancer VHL 1(3), 26 (2014)
Warren, A.Y., Harrison, D.: WHO/ISUP classification, grading and pathological staging of renal cell carcinoma: standards and controversies. World J. Urol. 36, 1913–1926 (2018)
Rees, M., Tekkis, P.P., Welsh, F.K., O’rourke, T., John, T.G.: Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann. Surg. 247(1), 125–135 (2008)
Zhu, M., Ren, B., Richards, R., Suriawinata, M., Tomita, N., Hassanpour, S.: Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Sci. Rep. 11(1), 1–9 (2021)
Abdeltawab, H.A., Khalifa, F.A., Ghazal, M.A., Cheng, L., El-Baz, A.S., Gondim, D.D.: A deep learning framework for automated classification of histopathological kidney whole-slide images. J. Pathol. Inform. 13, 100093 (2022)
Abu Haeyeh, Y., Ghazal, M., El-Baz, A., Talaat, I.M.: Development and evaluation of a novel deep-learning-based framework for the classification of renal histopathology images. Bioengineering 9(9), 423 (2022)
Fenstermaker, M., Tomlins, S.A., Singh, K., Wiens, J., Morgan, T.M.: Development and validation of a deep-learning model to assist with renal cell carcinoma histopathologic interpretation. Urology 144, 152–157 (2020)
Han, S., Hwang, S.I., Lee, H.J.: The classification of renal cancer in 3-phase CT images using a deep learning method. J. Digit. Imaging 32(4), 638–643 (2019)
Xi, I.L., et al.: Deep learning to distinguish benign from malignant renal lesions based on routine MR ImagingDeep learning for characterization of renal lesions. Clin. Cancer Res. 26(8), 1944–1952 (2020)
Baghdadi, A., et al.: Automated differentiation of benign renal oncocytoma and chromophobe renal cell carcinoma on computed tomography using deep learning. BJU Int. 125(4), 553–560 (2020)
Nikpanah, M., et al.: A deep-learning based artificial intelligence (AI) approach for differentiation of clear cell renal cell carcinoma from oncocytoma on multi-phasic MRI. Clin. Imaging 77, 291–298 (2021)
Zhou, L., Zhang, Z., Chen, Y.-C., Zhao, Z.-Y., Yin, X.-D., Jiang, H.-B.: A deep learning-based radiomics model for differentiating benign and malignant renal tumors. Transl. Oncol. 12(2), 292–300 (2019)
Hadjiyski, N.: Kidney cancer staging: deep learning neural network based approach. In: 2020 International Conference on e-Health and Bioengineering (EHB), pp. 1–4. IEEE (2020)
Hussain, M.A., Hamarneh, G., Garbi, R.: Renal cell carcinoma staging with learnable image histogram-based deep neural network. In: Suk, H.-I., Liu, M., Yan, P., Lian, C. (eds.) MLMI 2019. LNCS, vol. 11861, pp. 533–540. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32692-0_61
Delahunt, B., Cheville, J.C., et al.: The international society of urological pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am. J. Surg. Pathol. 37, 1490–1504 (2013)
Tan, M., Le, Q.V.: EfficientNet: rethinking model scaling for convolutional neural networks (2019)
Tian, K., et al.: Automated clear cell renal carcinoma grade classification with prognostic significance. PLoS ONE 14(10), e0222641 (2019)
Yeh, F.-C., Parwani, A.V., Pantanowitz, L., Ho, C.: Automated grading of renal cell carcinoma using whole slide imaging. J. Pathol. Inform. 5(1), 23 (2014)
Sun, X., et al.: Prediction of ISUP grading of clear cell renal cell carcinoma using support vector machine model based on CT images. Medicine 98(14) (2019)
Cui, E., et al.: Predicting the ISUP grade of clear cell renal cell carcinoma with multiparametric MR and multiphase CT radiomics. Eur. Radiol. 30, 2912–2921 (2020)
Zhao, Y., et al.: Deep learning based on MRI for differentiation of low- and high-grade in low-stage renal cell carcinoma. J. Magn. Reson. Imaging 52(5), 1542–1549 (2020)
Heller, N., et al.: The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: results of the kits19 challenge. Med. Image Anal. 101821 (2020)
Heller, N., et al.: The kits19 challenge data: 300 kidney tumor cases with clinical context, CT semantic segmentations, and surgical outcomes (2019)
Zhao, H., Li, H., Cheng, L.: Chapter 14 - data augmentation for medical image analysis. In: Burgos, N., Svoboda, D. (eds.) Biomedical Image Synthesis and Simulation. The MICCAI Society book Series, pp. 279–302. Academic Press (2022)
Chawla, N.V., Bowyer, K.W., Hall, L.O., Kegelmeyer, W.P.: SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002)
Shahinfar, S., Meek, P., Falzon, G.: “How many images do i need?” Understanding how sample size per class affects deep learning model performance metrics for balanced designs in autonomous wildlife monitoring. Ecol. Inform. 57, 101085 (2020)
Pérez-García, F., Sparks, R., Ourselin, S.: TorchIO: a python library for efficient loading, pre-processing, augmentation and patch-based sampling of medical images in deep learning. Comput. Methods Programs Biomed. 208, 106236 (2021)
Akar, E., Kara, S., Akdemir, H., Kiriş, A.: Fractal analysis of MR images in patients with chiari malformation: the importance of pre-processing. Biomed. Signal Process. Control 31, 63–70 (2017)
Alom, M.Z., Yakopcic, C., Hasan, M., Taha, T.M., Asari, V.K.: Recurrent residual U-Net for medical image segmentation. J. Med. Imaging (Bellingham) 6, 014006 (2019)
Vankdothu, R., Hameed, M.A.: Brain tumor MRI images identification and classification based on the recurrent convolutional neural network. Meas. Sens. 100412 (2022)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization (2014)
Boone, L., et al.: ROOD-MRI: benchmarking the robustness of deep learning segmentation models to out-of-distribution and corrupted data in MRI (2022)
Krishna, S.T., Kalluri, H.K.: Deep learning and transfer learning approaches for image classification (2019)
Shaha, M., Pawar, M.: Transfer learning for image classification. In: 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), pp. 656–660 (2018)
Hussain, M., Bird, J.J., Faria, D.R.: A study on CNN transfer learning for image classification. In: Lotfi, A., Bouchachia, H., Gegov, A., Langensiepen, C., McGinnity, M. (eds.) UKCI 2018. AISC, vol. 840, pp. 191–202. Springer, Cham (2019). https://doi.org/10.1007/978-3-319-97982-3_16
Yang, A., Yang, X., Wu, W., Liu, H., Zhuansun, Y.: Research on feature extraction of tumor image based on convolutional neural network. IEEE Access 7, 24204–24213 (2019)
Acknowledgement
This research was funded by the research council Norway and ICT:5G-HEART. We thank our colleagues Davit Aghayan and Egidijus Pelanis from Intervention Center, Oslo University Hospital, who provided insight and expertise in medical images and clinical aspects that greatly assisted the research. We gratitude Piotr Bialecki, Senior Engineering Manager of the PyTorch Team at NVIDIA, for assistance with technical parts of the training model with deep neural networks; and Håvard Kvamme, previous Ph.D. student of the University of Oslo in the faculty of Mathematics, for his comments and showing the actual path of the research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 ICST Institute for Computer Sciences, Social Informatics and Telecommunications Engineering
About this paper
Cite this paper
Mahootiha, M., Qadir, H.A., Bergsland, J., Balasingham, I. (2023). Classification of Kidney Tumor Grading on Preoperative Computed Tomography Scans. In: Tsanas, A., Triantafyllidis, A. (eds) Pervasive Computing Technologies for Healthcare. PH 2022. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering, vol 488. Springer, Cham. https://doi.org/10.1007/978-3-031-34586-9_6
Download citation
DOI: https://doi.org/10.1007/978-3-031-34586-9_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-34585-2
Online ISBN: 978-3-031-34586-9
eBook Packages: Computer ScienceComputer Science (R0)