Abstract
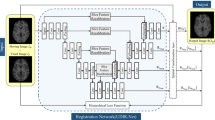
This paper presents a novel predictive model, MetaMorph, for metamorphic registration of images with appearance changes (i.e., caused by brain tumors). In contrast to previous learning-based registration methods that have little or no control over appearance-changes, our model introduces a new regularization that can effectively suppress the negative effects of appearance changing areas. In particular, we develop a piecewise regularization on the tangent space of diffeomorphic transformations (also known as initial velocity fields) via learned segmentation maps of abnormal regions. The geometric transformation and appearance changes are treated as joint tasks that are mutually beneficial. Our model MetaMorph is more robust and accurate when searching for an optimal registration solution under the guidance of segmentation, which in turn improves the segmentation performance by providing appropriately augmented training labels. We validate MetaMorph on real 3D human brain tumor magnetic resonance imaging (MRI) scans. Experimental results show that our model outperforms the state-of-the-art learning-based registration models. The proposed MetaMorph has great potential in various image-guided clinical interventions, e.g., real-time image-guided navigation systems for tumor removal surgery.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Alom, M.Z., Hasan, M., Yakopcic, C., Taha, T.M., Asari, V.K.: Recurrent residual convolutional neural network based on U-Net (R2U-Net) for medical image segmentation. arXiv preprint arXiv:1802.06955 (2018)
Arnold, V.: Sur la géométrie différentielle des groupes de lie de dimension infinie et ses applications à l’hydrodynamique des fluides parfaits. In: Annales de l’institut Fourier, vol. 16, pp. 319–361 (1966)
Arsigny, V., Commowick, O., Pennec, X., Ayache, N.: A log-Euclidean framework for statistics on diffeomorphisms. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI 2006. LNCS, vol. 4190, pp. 924–931. Springer, Heidelberg (2006). https://doi.org/10.1007/11866565_113
Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C.: Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12(1), 26–41 (2008)
Baid, U., et al.: The RSNA-ASNR-MICCAI brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:2107.02314 (2021)
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38(8), 1788–1800 (2019)
Beg, M.F., Miller, M.I., Trouvé, A., Younes, L.: Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int. J. Comput. Vis. 61(2), 139–157 (2005)
Bône, A., Vernhet, P., Colliot, O., Durrleman, S.: Learning joint shape and appearance representations with metamorphic auto-encoders. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12261, pp. 202–211. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59710-8_20
Chen, J., et al.: TransUNet: transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
Chen, J., Frey, E.C., He, Y., Segars, W.P., Li, Y., Du, Y.: TransMorph: transformer for unsupervised medical image registration. Med. Image Anal. 82, 102615 (2022)
Chen, M., Kanade, T., Pomerleau, D., Rowley, H.A.: Anomaly detection through registration. Pattern Recogn. 32(1), 113–128 (1999)
Chung, A.C.S., Wells, W.M., Norbash, A., Grimson, W.E.L.: Multi-modal image registration by minimising Kullback-Leibler distance. In: Dohi, T., Kikinis, R. (eds.) MICCAI 2002. LNCS, vol. 2489, pp. 525–532. Springer, Heidelberg (2002). https://doi.org/10.1007/3-540-45787-9_66
Dice, L.R.: Measures of the amount of ecologic association between species. Ecology 26(3), 297–302 (1945)
François, A., Gori, P., Glaunès, J.: Metamorphic image registration using a semi-Lagrangian scheme. In: Nielsen, F., Barbaresco, F. (eds.) GSI 2021. LNCS, vol. 12829, pp. 781–788. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-80209-7_84
Hatamizadeh, A., et al.: UNETR: transformers for 3D medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 574–584 (2022)
Holm, D., Trouvé, A., Younes, L.: The Euler-Poincaré theory of metamorphosis. Q. Appl. Math. 67(4), 661–685 (2009)
Kim, B., Han, I., Ye, J.C.: DiffuseMorph: unsupervised deformable image registration along continuous trajectory using diffusion models. arXiv preprint arXiv:2112.05149 (2021)
Maes, F., Collignon, A., Vandermeulen, D., Marchal, G., Suetens, P.: Multimodality image registration by maximization of mutual information. IEEE Trans. Med. Imaging 16(2), 187–198 (1997)
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014)
Miller, M.I., Trouvé, A., Younes, L.: Geodesic shooting for computational anatomy. J. Math. Imaging Vis. 24(2), 209–228 (2006)
Niethammer, M., et al.: Geometric metamorphosis. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011. LNCS, vol. 6892, pp. 639–646. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-23629-7_78
Nocedal, J., Wright, S.J.: Numerical Optimization. Springer, Heidelberg (1999)
Patenaude, B., Smith, S.M., Kennedy, D.N., Jenkinson, M.: A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 56(3), 907–922 (2011)
Prastawa, M., Bullitt, E., Ho, S., Gerig, G.: A brain tumor segmentation framework based on outlier detection. Med. Image Anal. 8(3), 275–283 (2004)
Qin, C., Shi, B., Liao, R., Mansi, T., Rueckert, D., Kamen, A.: Unsupervised deformable registration for multi-modal images via disentangled representations. In: Chung, A.C.S., Gee, J.C., Yushkevich, P.A., Bao, S. (eds.) IPMI 2019. LNCS, vol. 11492, pp. 249–261. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-20351-1_19
Rao, A., Aljabar, P., Rueckert, D.: Hierarchical statistical shape analysis and prediction of sub-cortical brain structures. Med. Image Anal. 12(1), 55–68 (2008)
Riklin-Raviv, T., Van Leemput, K., Menze, B.H., Wells, W.M., III., Golland, P.: Segmentation of image ensembles via latent atlases. Med. Image Anal. 14(5), 654–665 (2010)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Vialard, F.X., Risser, L., Rueckert, D., Cotter, C.J.: Diffeomorphic 3D image registration via geodesic shooting using an efficient adjoint calculation. Int. J. Comput. Vis. 97(2), 229–241 (2012)
Wachinger, C., Golland, P.: Atlas-based under-segmentation. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8673, pp. 315–322. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10404-1_40
Wang, J., Zhang, M.: DeepFLASH: an efficient network for learning-based medical image registration. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4444–4452 (2020)
Wang, J., Zhang, M.: Bayesian atlas building with hierarchical priors for subject-specific regularization. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12904, pp. 76–86. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87202-1_8
Wang, J., Zhang, M.: Geo-SIC: learning deformable geometric shapes in deep image classifiers. In: The Conference on Neural Information Processing Systems (2022)
Wells, W.M., III., Viola, P., Atsumi, H., Nakajima, S., Kikinis, R.: Multi-modal volume registration by maximization of mutual information. Med. Image Anal. 1(1), 35–51 (1996)
Zhang, M., Wells, W.M., Golland, P.: Low-dimensional statistics of anatomical variability via compact representation of image deformations. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9902, pp. 166–173. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46726-9_20
Zhao, S., Wang, Y., Yang, Z., Cai, D.: Region mutual information loss for semantic segmentation. In: Advances in Neural Information Processing Systems, vol. 32 (2019)
Acknowledgments
This work was supported by NSF CAREER Grant 2239977.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, J., Xing, J., Druzgal, J., Wells, W.M., Zhang, M. (2023). MetaMorph: Learning Metamorphic Image Transformation with Appearance Changes. In: Frangi, A., de Bruijne, M., Wassermann, D., Navab, N. (eds) Information Processing in Medical Imaging. IPMI 2023. Lecture Notes in Computer Science, vol 13939. Springer, Cham. https://doi.org/10.1007/978-3-031-34048-2_44
Download citation
DOI: https://doi.org/10.1007/978-3-031-34048-2_44
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-34047-5
Online ISBN: 978-3-031-34048-2
eBook Packages: Computer ScienceComputer Science (R0)