Abstract
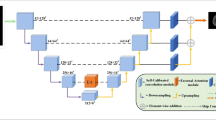
Brain tumor segmentation by computer computing is still an exciting challenge. UNet architecture has been widely used for medical image segmentation with several modifications. Attention blocks have been used to modify skip connections on the UNet architecture and result in improved performance. In this study, we propose the development of UNet for brain tumor image segmentation by modifying its contraction and expansion block by adding Attention, adding multiple atrous convolutions, and adding a residual pathway that we call Multiple Atrous convolutions Attention Block (MAAB). The expansion part is also added with the formation of pyramid features taken from each level to produce the final segmentation output. The architecture is trained using patches and batch 2 to save GPU memory usage. Online validation of the segmentation results from the BraTS 2021 validation dataset resulted in dice performance of 78.02, 80.73, and 89.07 for ET, TC, and WT. These results indicate that the proposed architecture is promising for further development.
Supported by Ministry of Education and Culture, Indonesia.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
References
Aghalari, M., Aghagolzadeh, A., Ezoji, M.: Brain tumor image segmentation via asymmetric/symmetric UNet based on two-pathway-residual blocks. Biomed. Signal Process. Control 69, 102841 (2021). https://doi.org/10.1016/j.bspc.2021.102841
Akbar, A.S., Fatichah, C., Suciati, N.: Simple myunet3d for brats segmentation. In: 2020 4th International Conference on Informatics and Computational Sciences (ICICoS), pp. 1–6 (2020). https://doi.org/10.1109/ICICoS51170.2020.9299072
Akbar, A.S., Fatichah, C., Suciati, N.: Modified mobilenet for patient survival prediction. In: Crimi, A., Bakas, S. (eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 374–387. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-72087-2_33
Baid, U., Ghodasara, S., Bilello, M., et al.: The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification, July 2021. http://arxiv.org/abs/2107.02314
Bakas, S., Akbari, H., Sotiras, A., et al.: Segmentation labels for the pre-operative scans of the tcga-gbm collection (2017). https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q, https://wiki.cancerimagingarchive.net/x/KoZyAQ
Bakas, S., Akbari, H., Sotiras, A., et al.: Segmentation labels for the pre-operative scans of the tcga-lgg collection (2017). https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF, https://wiki.cancerimagingarchive.net/x/LIZyAQ
Bakas, S., Akbari, H., Sotiras, A., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data 4(1), September 2017. https://doi.org/10.1038/sdata.2017.117
Chang, J., Zhang, L., Gu, N., et al.: A mix-pooling CNN architecture with FCRF for brain tumor segmentation. J. Visual Commun. Image Representation 58, 316–322 (2019). https://doi.org/10.1016/j.jvcir.2018.11.047
Kabir Anaraki, A., Ayati, M., Kazemi, F.: Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 39(1), 63–74 (2019). https://doi.org/10.1016/j.bbe.2018.10.004
Lin, T.Y., Dollár, P., Girshick, R., et al.: Feature pyramid networks for object detection. In: Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. vol. 2017-Janua, pp. 936–944. IEEE, July 2017. https://doi.org/10.1109/CVPR.2017.106. http://ieeexplore.ieee.org/document/8099589/
Liu, L., Wu, F.X., Wang, J.: Efficient multi-kernel DCNN with pixel dropout for stroke MRI segmentation. Neurocomputing 350, 117–127 (2019). https://doi.org/10.1016/j.neucom.2019.03.049
Menze, B.H., Jakab, A., Bauer, S., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). https://doi.org/10.1109/tmi.2014.2377694
Moradi, S., Oghli, M.G., Alizadehasl, A., et al.: MFP-Unet: a novel deep learning based approach for left ventricle segmentation in echocardiography. Phys. Medica 67, 58–69 (2019). https://doi.org/10.1016/J.EJMP.2019.10.001. https://www.sciencedirect.com/science/article/pii/S1120179719304508
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Schlemper, J., Oktay, O., Schaap, M., et al.: Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 53, 197–207 (2019). https://doi.org/10.1016/j.media.2019.01.012https://www.sciencedirect.com/science/article/pii/S1361841518306133
Srivastava, N., Hinton, G., Krizhevsky, A., et al.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15(1), 1929–1958 (2014)
S. V. and I. G.: Encoder enhanced atrous (EEA) unet architecture for retinal blood vessel segmentation. Cogn. Syst. Res. 67, 84–95 (2021). https://doi.org/10.1016/j.cogsys.2021.01.003
Wang, J., Gao, J., Ren, J., et al.: DFP-ResUNet: convolutional neural network with a dilated convolutional feature pyramid for multimodal brain tumor segmentation. Comput. Methods Programs Biomed., 106208, May 2021. https://doi.org/10.1016/j.cmpb.2021.106208.https://linkinghub.elsevier.com/retrieve/pii/S0169260721002820
Xie, H., Yang, D., Sun, N., et al.: Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recogn. 85, 109–119 (2019). https://doi.org/10.1016/j.patcog.2018.07.031
Yang, T., Song, J., Li, L.: A deep learning model integrating SK-TPCNN and random forests for brain tumor segmentation in MRI. Biocybern. Biomed. Eng. 39(3), 613–623 (2019). https://doi.org/10.1016/J.BBE.2019.06.003. https://www.sciencedirect.com/science/article/pii/S0208521618303292
Zhou, Z., He, Z., Jia, Y.: AFPNet: a 3D fully convolutional neural network with atrous-convolution feature pyramid for brain tumor segmentation via MRI images. Neurocomputing 402, 235–244 (2020). https://doi.org/10.1016/j.neucom.2020.03.097. https://www.sciencedirect.com/science/article/pii/S0925231220304847
Zhou, Z., He, Z., Shi, M., et al.: 3D dense connectivity network with atrous convolutional feature pyramid for brain tumor segmentation in magnetic resonance imaging of human heads. Comput. Biol. Med. 121, 103766 (2020). https://doi.org/10.1016/j.compbiomed.2020.103766
Acknowledgements
This work was supported by the Ministry of Education and Culture, Indonesia. We are deeply grateful for BPPDN (Beasiswa Pendidikan Pascasarjana Dalam Negeri) and PDD (Penelitian Disertasi Doktor) 2020–2021 Grant, which enabled this research could be done.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Akbar, A.S., Fatichah, C., Suciati, N. (2022). Unet3D with Multiple Atrous Convolutions Attention Block for Brain Tumor Segmentation. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2021. Lecture Notes in Computer Science, vol 12962. Springer, Cham. https://doi.org/10.1007/978-3-031-08999-2_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-08999-2_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-08998-5
Online ISBN: 978-3-031-08999-2
eBook Packages: Computer ScienceComputer Science (R0)