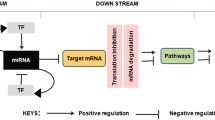
Abstract
MicroRNAs (miRNAs) provide a fundamental layer of regulation in cells. miRNAs act posttranscriptionally through complementary base-pairing with the 3′-UTR of a target mRNA, leading to mRNA degradation and translation arrest. The likelihood of forming a valid miRNA-target duplex within cells was computationally predicted and experimentally monitored. In human cells, the miRNA profiles determine their identity and physiology. Therefore, alterations in the composition of miRNAs signify many cancer types and chronic diseases. In this chapter, we introduce online functional tools and resources to facilitate miRNA research. We start by introducing currently available miRNA catalogs and miRNA-gateway portals for navigating among different miRNA-centric online resources. We then sketch several realistic challenges that may occur while investigating miRNA regulation in living cells. As a showcase, we demonstrate the utility of miRNAs and mRNAs expression databases that cover diverse human cells and tissues, including resources that report on genetic alterations affecting miRNA expression levels and alteration in binding capacity. Introducing tools linking miRNAs with transcription factor (TF) networks reveals miRNA regulation complexity within living cells. Finally, we concentrate on online resources that analyze miRNAs in human diseases and specifically in cancer. Altogether, we introduce contemporary, selected resources and online tools for studying miRNA regulation in cells and tissues and their utility in health and disease.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- CAGE:
-
Cap-based expression analysis
- ceRNA:
-
Competing endogenous RNA
- ChIP:
-
Chromatin immunoprecipitation
- circRNA:
-
Circular RNA
- CLASH:
-
Cross linking, ligation and sequencing of hybrids
- CLIP:
-
Cross-linking immunoprecipitation
- CNV:
-
Copy number variation
- DRV:
-
Disease-related variation
- FFL:
-
Feed-forward loop
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- GWAS:
-
Genome wide association study
- HTP:
-
High throughput
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- lncRNA:
-
Long non-coding RNAs
- LTP:
-
Low throughput
- MBS:
-
miRNA-binding sites
- miRNA:
-
microRNA
- ML:
-
Machine learning
- mRNA:
-
Messenger RNA
- MS:
-
Mass spectrometry
- MTI:
-
miRNA-target interaction
- RISC:
-
RNA-induced silencing complex
- RPM:
-
Reads per million
- Seq:
-
Sequencing
- smRNA:
-
Small RNA
- SNV:
-
Single nucleotide variation
- SVM:
-
Support vector machine
- TCGA:
-
The Cancer Genome Atlas
- TF:
-
Transcription factor
- TFBS:
-
TF binding sites
- TSS:
-
Transcription start sites
- UTR:
-
Untranslated region
References
Abugessaisa I, Ramilowski JA, Lizio M, Severin J, Hasegawa A, Harshbarger J, Kondo A, Noguchi S, Yip CW, Ooi JLC (2021) FANTOM enters 20th year: expansion of transcriptomic atlases and functional annotation of non-coding RNAs. Nucleic Acids Res 49:D892–D898
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. elife 4
Aghaee-Bakhtiari SH, Arefian E, Lau P (2018) miRandb: a resource of online services for miRNA research. Brief Bioinform 19:254–262
Ahadi A, Sablok G, Hutvagner G (2017) miRTar2GO: a novel rule-based model learning method for cell line specific microRNA target prediction that integrates Ago2 CLIP-Seq and validated microRNA–target interaction data. Nucleic Acids Res 45:e42–e42
Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, García Girón C, Hourlier T (2016) The Ensembl gene annotation system. Database 2016
Akhtar MM, Micolucci L, Islam MS, Olivieri F, Procopio AD (2016) Bioinformatic tools for microRNA dissection. Nucleic Acids Res 44:24–44
Alexiou P, Vergoulis T, Gleditzsch M, Prekas G, Dalamagas T, Megraw M, Grosse I, Sellis T, Hatzigeorgiou AG (2010) miRGen 2.0: a database of microRNA genomic information and regulation. Nucleic Acids Res 38:D137–D141
Alvarez-Garcia I, Miska EA (2005) MicroRNA functions in animal development and human disease. Development 132:4653–4662
Andres-Leon E, Gonzalez Pena D, Gomez-Lopez G, Pisano DG (2015) miRGate: a curated database of human, mouse and rat miRNA-mRNA targets. Database (Oxford) bav035
Aparicio-Puerta E, Lebron R, Rueda A, Gomez-Martin C, Giannoukakos S, Jaspez D, Medina JM, Zubkovic A, Jurak I, Fromm B, Marchal JA, Oliver J, Hackenberg M (2019) smRNAbench and smRNAtoolbox 2019: intuitive fast small RNA profiling and differential expression. Nucleic Acids Res 47:W530–W535
Backes C, Kehl T, Stöckel D, Fehlmann T, Schneider L, Meese E, Lenhof H-P, Keller A (2016) miRPathDB: a new dictionary on microRNAs and target pathways. Nucleic Acids Res gkw926
Backes C, Fehlmann T, Kern F, Kehl T, Lenhof H-P, Meese E, Keller A (2018) miRCarta: a central repository for collecting miRNA candidates. Nucleic Acids Res 46:D160–D167
Balaga O, Friedman Y, Linial M (2012) Toward a combinatorial nature of microRNA regulation in human cells. Nucleic Acids Res 40:9404–9416
Bhartiya D, Laddha SV, Mukhopadhyay A, Scaria V (2011) miRvar: a comprehensive database for genomic variations in microRNAs. Hum Mutat 32:E2226–E2245
Bhattacharya A, Cui Y (2016) SomamiR 2.0: a database of cancer somatic mutations altering microRNA–ceRNA interactions. Nucleic Acids Res 44:D1005–D1010
Bhattacharya A, Ziebarth JD, Cui Y (2014) PolymiRTS database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res 42:D86–D91
Biggar KK, Storey KB (2015) Insight into post-transcriptional gene regulation: stress-responsive microRNAs and their role in the environmental stress survival of tolerant animals. J Exp Biol 218:1281–1289
Bruno AE, Li L, Kalabus JL, Pan Y, Yu A, Hu Z (2012) miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC Genomics 13:1–7
Cai Y, Yu X, Hu S, Yu J (2009) A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 7:147–154
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48:D127–D131
Chen L, Heikkinen L, Wang C, Yang Y, Knott KE, Wong G (2018a) miRToolsGallery: a tag-based and rankable microRNA bioinformatics resources database portal. Database (Oxford)
Chen X, Wang C-C, Yin J, You Z-H (2018b) Novel human miRNA-disease association inference based on random forest. Molecular Therapy-Nucleic Acids 13:568–579
Chen X, Yin J, Qu J, Huang L (2018c) MDHGI: matrix decomposition and heterogeneous graph inference for miRNA-disease association prediction. PLoS Comput Biol 14:e1006418
Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G (2019) Trends in the development of miRNA bioinformatics tools. Brief Bioinform 20:1836–1852
Cho S, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, Choi I, Chang H, Ryu D, Lee B, Kim VN, Kim W, Lee S (2013) MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res 41:D252–D257
Chou CH, Lin FM, Chou MT, Hsu SD, Chang TH, Weng SL, Shrestha S, Hsiao CC, Hung JH, Huang HD (2013) A computational approach for identifying microRNA-target interactions using high-throughput CLIP and PAR-CLIP sequencing. BMC Genomics 14(Suppl 1):S2
Das SS, Saha P, Chakravorty N (2018) miRwayDB: a database for experimentally validated microRNA-pathway associations in pathophysiological conditions. Database (Oxford)
Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK (2018) The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46:D794–D801
Djuranovic S, Nahvi A, Green R (2011) A parsimonious model for gene regulation by miRNAs. Science 331:550–553
Dweep H, Gretz N (2015) miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 12:697
Friard O, Re A, Taverna D, De Bortoli M, Cora D (2010) CircuitsDB: a database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse. BMC Bioinformatics 11:435
Friedman Y, Karsenty S, Linial M (2014) miRror-suite: decoding coordinated regulation by microRNAs. Database (Oxford)
Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ (2015) A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet 49:213–242
Fromm B, Domanska D, Hoye E, Ovchinnikov V, Kang W, Aparicio-Puerta E, Johansen M, Flatmark K, Mathelier A, Hovig E, Hackenberg M, Friedlander MR, Peterson KJ (2020) MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res 48:D1172
Fu G, Brkić J, Hayder H, Peng C (2013) MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci 14:5519–5544
Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A (2010) The UCSC genome browser database: update 2011. Nucleic Acids Res 39:D876–D882
Gebert LF, MacRae IJ (2019) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 20:21–37
Gerlach D, Kriventseva EV, Rahman N, Vejnar CE, Zdobnov EM (2009) miROrtho: computational survey of microRNA genes. Nucleic Acids Res 37:D111–D117
Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, Barretina J, Gelfand ET, Bielski CM, Li H (2019) Next-generation characterization of the cancer cell line encyclopedia. Nature 569:503–508
Glogovitis I, Yahubyan G, Wurdinger T, Koppers-Lalic D, Baev V (2020) isomiRs-hidden soldiers in the miRNA regulatory Army, and how to find them? Biomol Ther 11
Gomes CPDC, Cho J-H, Hood LE, Franco OL, Pereira RWD, Wang K (2013) A review of computational tools in microRNA discovery. Front Genet 4:81
Griffiths-Jones S (2004) The microRNA registry. Nucleic Acids Res 32:D109–D111
Gu C, Liao B, Li X, Li K (2016) Network consistency projection for human miRNA-disease associations inference. Sci Rep 6:1–10
Guo Z, Maki M, Ding R, Yang Y, Xiong L (2014) Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep 4:1–9
Hamed M, Spaniol C, Nazarieh M, Helms V (2015) TFmiR: a web server for constructing and analyzing disease-specific transcription factor and miRNA co-regulatory networks. Nucleic Acids Res 43:W283–W288
Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li J, Zhou Y, Cui Q (2019) HMDD v3. 0: a database for experimentally supported human microRNA–disease associations. Nucleic Acids Res 47:D1013–D1017
Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, Tang Y, Chen YG, Jin CN, Yu Y, Xu JT, Li YM, Cai XX, Zhou ZY, Chen XH, Pei YY, Hu L, Su JJ, Cui SD, Wang F, Xie YY, Ding SY, Luo MF, Chou CH, Chang NW, Chen KW, Cheng YH, Wan XH, Hsu WL, Lee TY, Wei FX, Huang HD (2020) miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res 48:D148–D154
Ison J, Ienasescu H, Chmura P, Rydza E, Ménager H, Kalaš M, Schwämmle V, Grüning B, Beard N, Lopez R (2019) The bio. tools registry of software tools and data resources for the life sciences. Genome Biol 20:1–4
Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y (2009) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 37:D98–D104
Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI (2018) Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46:D335–D342
Kamanu TK, Radovanovic A, Archer JA, Bajic VB (2013) Exploration of miRNA families for hypotheses generation. Sci Rep 3:1–8
Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG (2018) DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 46:D239–D245
Kehl T, Kern F, Backes C, Fehlmann T, Stockel D, Meese E, Lenhof HP, Keller A (2020) miRPathDB 2.0: a novel release of the miRNA pathway dictionary database. Nucleic Acids Res 48:D142–D147
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162
Kuenne C, Preussner J, Herzog M, Braun T, Looso M (2014) MIRPIPE: quantification of microRNAs in niche model organisms. Bioinformatics 30:3412–3413
Kyrollos DG, Reid B, Dick K, Green JR (2020) RPmirDIP: reciprocal perspective improves targeting prediction. Sci Rep 10:11770
Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H (2014) starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42:D92–D97
Liu CJ, Fu X, Xia M, Zhang Q, Gu Z, Guo AY (2021) miRNASNP-v3: a comprehensive database for SNPs and disease-related variations in miRNAs and miRNA targets. Nucleic Acids Res 49:D1276–D1281
Loher P, Rigoutsos I (2012) Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 28:3322–3323
Lukasik A, Zielenkiewicz P (2019) An overview of miRNA and miRNA target analysis tools. Methods Mol Biol 1932:65–87
Lukasik A, Wojcikowski M, Zielenkiewicz P (2016) Tools4miRs - one place to gather all the tools for miRNA analysis. Bioinformatics 32:2722–2724
Mahlab-Aviv S, Linial N, Linial M (2019) A cell-based probabilistic approach unveils the concerted action of miRNAs. PLoS Comput Biol 15:e1007204
Maselli V, Di Bernardo D, Banfi S (2008) CoGemiR: a comparative genomics microRNA database. BMC Genomics 9:457
Mendes ND, Freitas AT, Sagot MF (2009) Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res 37:2419–2433
Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S (2017) Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform 18:780–788
Monga I, Kumar M (2019) Computational resources for prediction and analysis of functional miRNA and their targetome. Methods Mol Biol 1912:215–250
Morel L, Regan M, Higashimori H, Ng SK, Esau C, Vidensky S, Rothstein J, Yang Y (2013) Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J Biol Chem 288:7105–7116
Müller S, Rycak L, Afonso-Grunz F, Winter P, Zawada AM, Damrath E, Scheider J, Schmäh J, Koch I, Kahl G (2014) APADB: a database for alternative polyadenylation and microRNA regulation events. Database
Naamati G, Friedman Y, Balaga O, Linial M (2012) Susceptibility of the human pathways graphs to fragmentation by small sets of microRNAs. Bioinformatics 28:983–990
Panwar B, Omenn GS, Guan Y (2017) miRmine: a database of human miRNA expression profiles. Bioinformatics 33:1554–1560
Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG (2013) DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41:W169–W173
Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T (2016) DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res 44:D231–D238
Perdikopanis N, Georgakilas GK, Grigoriadis D, Pierros V, Kavakiotis I, Alexiou P, Hatzigeorgiou A (2021) DIANA-miRGen v4: indexing promoters and regulators for more than 1500 microRNAs. Nucleic Acids Res 49:D151–D159
Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB (2014) Common features of microRNA target prediction tools. Front Genet 5
Preusse M, Theis FJ, Mueller NS (2016) miTALOS v2: analyzing tissue specific microRNA function. PLoS One 11:e0151771
Re A, Caselle M, Bussolino F (2017) MicroRNA-mediated regulatory circuits: outlook and perspectives. Phys Biol 14:045001
Riffo-Campos AL, Riquelme I, Brebi-Mieville P (2016) Tools for sequence-based miRNA target prediction: what to choose? Int J Mol Sci 17
Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, Mahaffey S, Rossi S, Calin GA, Bemis L, Theodorescu D (2014) The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res 42:e133
Rueda A, Barturen G, Lebron R, Gomez-Martin C, Alganza A, Oliver JL, Hackenberg M (2015) smRNAtoolbox: an integrated collection of small RNA research tools. Nucleic Acids Res 43:W467–W473
Russo F, Di Bella S, Vannini F, Berti G, Scoyni F, Cook HV, Santos A, Nigita G, Bonnici V, Lagana A, Geraci F, Pulvirenti A, Giugno R, De Masi F, Belling K, Jensen LJ, Brunak S, Pellegrini M, Ferro A (2018) miRandola 2017: a curated knowledge base of non-invasive biomarkers. Nucleic Acids Res 46:D354–D359
Sarver AL, Sarver AE, Yuan C, Subramanian S (2018) OMCD: Oncomir cancer database. BMC Cancer 18:1–6
Sato F, Tsuchiya S, Meltzer SJ, Shimizu K (2011) MicroRNAs and epigenetics. FEBS J 278:1598–1609
Schmitz U, Wolkenhauer O (2013) Web resources for microRNA research. Adv Exp Med Biol 774:225–250
Shaker F, Nikravesh A, Arezumand R, Aghaee-Bakhtiari SH (2020) Web-based tools for miRNA studies analysis. Comput Biol Med 127:104060
Shirdel EA, Xie W, Mak TW, Jurisica I (2011) NAViGaTing the micronome—using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One 6:e17429
Shuang C, Maozu G, Chunyu W, Xiaoyan L, Yang L, Xuejian W (2016) MiRTDL: a deep learning approach for miRNA target prediction. IEEE/ACM Trans Comput Biol Bioinform 13:1161–1169
Shukla V, Varghese VK, Kabekkodu SP, Mallya S, Satyamoorthy K (2017) A compilation of web-based research tools for miRNA analysis. Brief Funct Genomics 16:249–273
Solomon J, Kern F, Fehlmann T, Meese E, Keller A (2020) HumiR: web services, tools and databases for exploring human microRNA data. Biomolecules 10
Sticht C, De La Torre C, Parveen A, Gretz N (2018) miRWalk: an online resource for prediction of microRNA binding sites. PLoS One 13:e0206239
The RNAcentral Consortium, Petrov AI, Kay SJE, Kalvari I, Howe KL, Gray KA, Bruford EA, Kersey PJ, Cochrane G, Finn RD, Bateman A, Kozomara A, Griffiths-Jones S, Frankish A, Zwieb CW, Lau BY, Williams KP, Chan PP, Lowe TM, Cannone JJ, Gutell R, Machnicka MA, Bujnicki JM, Yoshihama M, Kenmochi N, Chai B, Cole JR, Szymanski M, Karlowski WM, Wood V, Huala E, Berardini TZ, Zhao Y, Chen R, Zhu W, Paraskevopoulou MD, Vlachos IS, Hatzigeorgiou AG, Ma L, Zhang Z, Puetz J, Stadler PF, McDonald D, Basu S, Fey P, Engel SR, Cherry JM, Volders P-J, Mestdagh P, Wower J, Clark MB, Quek XC, Dinger ME (2017) RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res 45:D128–D134
Thomson DW, Bracken CP, Goodall GJ (2011) Experimental strategies for microRNA target identification. Nucleic Acids Res 39:6845–6853
Tokar T, Pastrello C, Rossos AE, Abovsky M, Hauschild A-C, Tsay M, Lu R, Jurisica I (2018) mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res 46:D360–D370
Tong Y, Ru B, Zhang J (2018) miRNACancerMAP: an integrative web server inferring miRNA regulation network for cancer. Bioinformatics 34:3211–3213
Tong Z, Cui Q, Wang J, Zhou Y (2019) TransmiR v2.0: an updated transcription factor-microRNA regulation database. Nucleic Acids Res 47:D253–D258
Van Peer G, Lefever S, Anckaert J, Beckers A, Rihani A, Van Goethem A, Volders PJ, Zeka F, Ongenaert M, Mestdagh P, Vandesompele J (2014) miRBase tracker: keeping track of microRNA annotation changes. Database (Oxford)
Vejnar CE, Blum M, Zdobnov EM (2013) miRmap web: comprehensive microRNA target prediction online. Nucleic Acids Res 41:W165–W168
Wang P, Zhi H, Zhang Y, Liu Y, Zhang J, Gao Y, Guo M, Ning S, Li X (2015) miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database
Wienholds E, Plasterk RH (2005) MicroRNA function in animal development. FEBS Lett 579:5911–5922
Wong NW, Chen Y, Chen S, Wang X (2018) OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 34:713–715
Wu W-S, Tu B-W, Chen T-T, Hou S-W, Tseng JT (2017) CSmiRTar: condition-specific microRNA targets database. PLoS One 12:e0181231
Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T (2009) miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37:D105–D110
Xie B, Ding Q, Han H, Wu D (2013) miRCancer: a microRNA–cancer association database constructed by text mining on literature. Bioinformatics 29:638–644
Yang J-H, Li J-H, Jiang S, Zhou H, Qu L-H (2013) ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res 41:D177–D187
Yang Z, Wu L, Wang A, Tang W, Zhao Y, Zhao H, Teschendorff AE (2017) dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Res 45:D812–D818
You Z-H, Huang Z-A, Zhu Z, Yan G-Y, Li Z-W, Wen Z, Chen X (2017) PBMDA: a novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput Biol 13:e1005455
Yue D, Liu H, Huang Y (2009) Survey of computational algorithms for MicroRNA target prediction. Curr Genomics 10:478–492
Zou Q, Li J, Hong Q, Lin Z, Wu Y, Shi H, Ju Y (2015) Prediction of microRNA-disease associations based on social network analysis methods. BioMed Res Int
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Blass, I., Zohar, K., Linial, M. (2022). Turning Data to Knowledge: Online Tools, Databases, and Resources in microRNA Research. In: Schmitz, U., Wolkenhauer, O., Vera-González, J. (eds) Systems Biology of MicroRNAs in Cancer. Advances in Experimental Medicine and Biology, vol 1385. Springer, Cham. https://doi.org/10.1007/978-3-031-08356-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-031-08356-3_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-08355-6
Online ISBN: 978-3-031-08356-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)