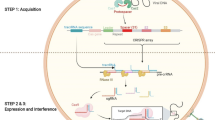
Abstract
At present, traditional biotechnology techniques have usually been exploited for medicinal plant breeding. However, recent biotechnology-based breeding techniques, i.e., genome editing diversify the platform to develop custom-designed medicinal plants. The sequence-specific nucleases of TALENs, ZFNs, and Cas are advance genome editing tools which produce user-defined plants. CRISPR/Cas-based genome editing is an emerging technique that utilizes artificially engineered nucleases for digesting DNA at targeted locations in the genome for high-throughput biotechnology-based breeding of valuable medicinal plants. Due to its wide application in gene mutagenesis, transcriptional regulation, high efficiency and easy manipulation, several plant-specific CRISPR/Cas9 vector systems have been designed. This methodology is based upon the type II adaptive immunity response of prokaryotes, which comprises of a CRISPR-associated (Cas)9 protein and an engineered sgRNA that specifically targets the nucleic acid sequence to induce selective mutagenesis. In this chapter, various CRISPR/Cas-based approaches are discussed with emphasis on CRISPR/Cas9 vector platforms, multiplex editing strategies, analysis methods for induced mutations, and its applications in medicinal plants. This chapter provides the advancements in genome editing technologies and their associated strategies, giving an insight on the limitations of CRISPR/Cas9 technique and its future advances to improve the quality of traditional medicinal herbs. This new system will further open new arena to manage synthetic biology of medicinal plants for industrial purposes and to investigate the function of gene to accelerate basic plant research.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Adli M (2018) The CRISPR tool kit for genome editing and beyond. Nat Commun 9:1911
Bahuguna RN, Joshi R, Singh G, Shukla A, Gupta R, Bains G (2011) Micropropagation and total alkaloid extraction of Indian snake root (Rauwolfia serpentina). Indian J Agric Sci 81:1124–1129
Bibikova M, Beumer K, Trautman JK, Carroll D (2003) Enhancing gene targeting with designed zinc finger nucleases. Science 300(5620):764
Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436
Chandrasegaran S, Carroll D (2016) Origins of programmable nucleases for genome engineering. J Mol Biol 428(5):963–989
Chen Z, Wu J, Ma Y, Wang P, Gu Z, Yang R (2018) Biosynthesis, metabolic regulation and bioactivity of phenolic acids in plant food materials. Food Sci 39(7):321–328
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823
Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S (2015) Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol 33(11):1159–1161
Denbow CJ, Lapins S, Dietz N, Scherer R, Nimchuk ZL, Okumoto S (2017) Gateway-compatible CRISPR-Cas9 vectors and a rapid detection by high-resolution melting curve analysis. Front Plant Sci 8:1171
Dhar MK, Koul A, Kaul S (2013) Farnesyl pyrophosphate synthase: a key enzyme in isoprenoid biosynthetic pathway and potential molecular target for drug development. New Biotechnol 30(2):114–123
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346:12580
Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33(18):5978–5990
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci 111(12):4632–4637
Feng S, Song W, Fu R, Zhang H, Xu A, Li J (2018) Application of the CRISPR/Cas9 system in Dioscorea zingiberensis. Plant Cell Tissue Organ Cult 135(1):133–141
Fuller KK, Chen S, Loros JJ, Dunlap JC (2015) Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell 14(11):1073–1080
Iqbal Z, Iqbal MS, Ahmad A, Memon AG, Ansari MI (2020) New prospects on the horizon: genome editing to engineer plants for desirable traits. Curr Plant Biol 20:100171
Ivanov IE, Wright AV, Cofsky JC, Aris KD, Doudna JA, Bryant Z (2020) Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc Natl Acad Sci 117(11):5853–5860
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821
Joshi R, Singh B, Chinnusamy V (2018) Genetically engineering cold stress-tolerant crops: approaches and challenges. In: Cold tolerance in plants. Springer, Cham, pp 179–195
Joshi R, Gupta BK, Pareek A, Singh MB, Singla-Pareek SL (2019) Functional genomics approach towards dissecting out abiotic stress tolerance trait in plants. In: Genetic enhancement of crops for tolerance to abiotic stress: mechanisms and approaches. Springer, Cham, pp 1–24
Joung JK, Sander JD (2013) TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14(1):49–55
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucl Acids Res 41(20): e188.
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93(3):1156–1160
Kui L, Chen H, Zhang W, He S, Xiong Z, Zhang Y, Yan L, Zhong C, He F, Chen J, Zeng P (2017) Building a genetic manipulation tool box for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front Plant Sci 7:2036
LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y (2018) Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93(2):377–386
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31(8):688–691
Li B, Cui G, Shen G, Zhan Z, Huang L, Chen J, Qi X (2017) Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci Rep 7(1):43320
Li R, Ba Y, Song Y, Cui J, Zhang X, Zhang D, Yuan Z, Yang L (2020) Rapid and sensitive screening and identification of CRISPR/Cas9 edited rice plants using quantitative real-time PCR coupled with high resolution melting analysis. Food Control 112:107088
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211
Liu J, Gunapati S, Mihelich NT, Stec AO, Michno JM, Stupar RM (2019) Genome editing in soybean with CRISPR/Cas9. In: Plant genome editing with CRISPR systems. Humana Press, New York, pp 217–234
Long L, Guo DD, Gao W, Yang WW, Hou LP, Ma XN, Miao YC, Botella JR, Song CP (2018) Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 14(1):85
Lowder L, Malzahn A, Qi Y (2017) Rapid construction of multiplexed CRISPR-Cas9 systems for plant genome editing. In: Plant pattern recognition receptors. Humana Press, New York, pp 291–307
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Ma X, Zhu Q, Chen Y, Liu YG (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant 9(7):961–974
Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat Plants 6(7):773–779
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol Plant 6(6):2008–2011
Moradpour M, Abdulah SN (2020) CRISPR/dC as9 platforms in plants: strategies and applications beyond genome editing. Plant Biotechnol J 18(1):32–44
Negritto MC (2010) Repairing double-strand DNA breaks. Nat Educ 3:26
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31(8):691–693
Niazian M (2019) Application of genetics and biotechnology for improving medicinal plants. Planta 249(4):953–973
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O (2014) Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156(5):935–949
Osakabe Y, Osakabe K (2015) Genome editing with engineered nucleases in plants. Plant Cell Physiol 56(3):389–400
Osakabe K, Saika H, Okuzaki A, Toki S (2012) Site-directed mutagenesis in higher plants. In: Shu QY, Forster BP, Nakagawa H (eds) Plant mutation breeding and biotechnology. CAB eBooks, Wallingford, pp 523–533
Pannunzio NR, Watanabe G, Lieber MR (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 293(27):10512–10523
Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2015) RNA-guided transcriptional regulation in planta via synthetic dC as9-based transcription factors. Plant Biotechnol J 13(4):578–589
Polstein LR, Gersbach CA (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11(3):198–200
Ren C, Liu Y, Guo Y, Duan W, Fan P, Li S, Liang Z (2021) Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic Res 8(1):52
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31(8):686–688
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507(7490):62–67
Terns MP, Terns RM (2011) CRISPR-based adaptive immune systems. Curr Opin Microbiol 14(3):321–327
Upadhyay J, Upadhyay G, Joshi R, Juyal V (2014) Effect of rhododendron flower juice on the bioavailability of amlodipine in rats. Int J Bioassays 3(2):1734–1737
Vats S, Kumawat S, Kumar V, Patil GB, Joshi T, Sonah H, Sharma TR, Deshmukh R (2019) Genome editing in plants: exploration of technological advancements and challenges. Cell 8(11):1386
Wang M, Mao Y, Lu Y, Wang Z, Tao X, Zhu JK (2018) Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems. J Intgr Plant Biol 60(8):626–631
Wani SH, Tripathi P, Zaid A, Challa GS, Kumar A, Kumar V, Upadhyay J, Joshi R, Bhatt M (2018) Transcriptional regulation of osmotic stress tolerance in wheat (Triticum aestivum L.). Plant Mol Biol 97(6):469–487
Xie X, Ma X, Liu YG (2019) Decoding sanger sequencing chromatograms from CRISPR-induced mutations. In: Plant genome editing with CRISPR systems. Humana Press, New York, pp 33–43
Yang L, Ding G, Lin H, Cheng H, Kong Y, Wei Y, Fang X, Liu R, Wang L, Chen X, Yang C (2013) Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS One 8(11):e80464
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/C as9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12(6):797–807
Zhou Z, Tan H, Li Q, Chen J, Gao S, Wang Y, Chen W, Zhang L (2018) CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 148:63–70
Zischewski J, Fischer R, Bortesi L (2017) Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol Adv 35(1):95–104
Acknowledgments
MP, KD and RJ gratefully acknowledge the director, CSIR-Institute of Himalayan Bioresource Technology, Palampur, for providing the facilities to carry out this work. CSIR support in the form of project MLP0201, MLP0165, MLP0170, and MLP0172 and agriculture department, Shimla in the form of project SSP-0125 and SSP-0126 for this study is highly acknowledged. This manuscript represents CSIR-IHBT communication no. 4891.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Patial, M., Devi, K., Joshi, R. (2022). CRISPR/Cas9-Mediated Targeted Mutagenesis in Medicinal Plants. In: Wani, S.H., Hensel, G. (eds) Genome Editing. Springer, Cham. https://doi.org/10.1007/978-3-031-08072-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-031-08072-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-08071-5
Online ISBN: 978-3-031-08072-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)