Abstract
The high incidence of cervical cancer in women has prompted the research of automatic screening methods. This work focuses on two of the steps present in such systems, more precisely, the identification of cervical lesions and their respective classification. The development of automatic methods for these tasks is associated with some shortcomings, such as acquiring sufficient and representative clinical data. These limitations are addressed through a hybrid pipeline based on a deep learning model (RetinaNet) for the detection of abnormal regions, combined with random forest and SVM classifiers for their categorization, and complemented by the use of domain knowledge in its design. Additionally, the nuclei in each detected region are segmented, providing a set of nuclei-specific features whose impact on the classification result is also studied. Each module is individually assessed in addition to the complete system, with the latter achieving a precision, recall and F1 score of 0.04, 0.20 and 0.07, respectively. Despite the low precision, the system demonstrates potential as an analysis support tool with the capability of increasing the overall sensitivity of the human examination process.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cervical cancer is the fourth most common type of cancer in women worldwide, despite being one of the more easily treatable types of cancer when detected early on [1]. Owing partially to this, there has been a significant research effort into effective screening methods over the last decades. The Papanicolau (or Pap) test is one of the most widely used, and it consists of analysing cells scraped from the squamocolumnar junction in the cervix [2] for malignant or pre-malignant signs. When performed manually, this is a very demanding task that requires highly specialised resources. Therefore, automated systems have gained a particular interest in more recent decades.
These computer-aided diagnosis (CAD) systems usually consider different steps in order to analyze each microscopic image from a sample: focus and adequacy assessment, region of interest (ROI) identification and respective classification [3]. Currently, there are still limitations associated with the last two stages, namely the acquisition of adequate clinical data in sufficient amounts.
In order to overcome these challenges, this work explores the combination of deep learning (DL) and conventional machine learning (ML) algorithms in hybrid pipelines for the detection and classification of abnormal regions in liquid-based cytology (LBC) images. While hybrid approaches have been investigated before in similar settings, most of these studies do not explicitly employ both types of algorithms for the detection and classification stages, using them instead for related tasks, such as feature extraction. Furthermore, the proposed system incorporates specific domain knowledge in its design, assessing the impact of clinically relevant features, such as those of the nuclei, in the algorithms’ discerning ability between different cervical lesions. In this manner, the present work provides a fresh perspective in terms of the architecture of CAD systems.
2 Related Work
With the technological advances in computer hardware and slide preparation techniques over the years, numerous approaches for the automatic analysis of cervical cytology images have been proposed [3]. When it comes to detect and isolate regions of interest in these images, the approaches more frequently encountered are based on conventional computer vision (CV) techniques aiming to segment an image’s cells and their inner structures.
The International Symposium on Biomedical Imaging (ISBI) challenges focus precisely on this issue, with some innovative works being produced within this scope. For example, the submission ranked first in the 2014 challenge by Ushizima, Bianchi and Carneiro [4] used a graph-based algorithm to identify cellular clumps (aggregates of partially overlapping cells), merging regions based on pixel adjacency and intensity similarity. As for the winners of the 2015 version of the challenge, Phoulady et al. [5] used iterative thresholding to detect cell’s nuclei. In [6], the authors also employed a classical approach to analyse Pap smear slides, namely using a sliding window strategy followed by the cells segmentation through a mini-batch k-means algorithm, which allows for the generation of 33 features that are later used for classification purposes.
Despite the major significance of the features that can be extracted from an adequately segmented image, this task is not easy to accomplish. Subsequently, deep learning techniques have emerged as alternatives for the detection and classification of abnormal regions. As an example, Du et al. [7] proposed a Faster R-CNN based system for the detection and classification of cervical cells from LBC samples, obtaining the best performance with a ResNet101 backbone when compared with other convolutional neural network (CNN) backbones. Zhou et al. [8] also performed several experiments for detecting abnormal cells in cervical pathology images with deep learning models, testing multiple architectures from which the RetinaNet achieved the highest average precision.
Regarding the classification of cervical cells, this task has had a bigger focus than their detection, partially because of the larger amount of available data directed towards it. Some of the aforementioned works [6,7,8] resort to deep neural networks for the classification of the cells or regions previously located. Similarly, Zhang et al. [9] and Kwon et al. [10] also used DL methods to tackle this task, more specifically through the use of custom networks, obtaining an accuracy of 98.2% and 84.5%, respectively. Ghoneim, Muhammad and Hossain [11] also performed several experiments with DL methods in which three separate networks were compared as feature extractors matched with three types of classifiers, with the best combination obtained being with a CaffeNet and an extreme learning machine. In the work of Su et al. [12], a more traditional approach used a C4.5 decision tree and a logistic regression algorithm to classify the cells, achieving results comparable to the deep learning approaches. Prum, Handayani and Boursier [13] also proposed a conventional system using a support vector machine (SVM) to classify the cells based on the histogram of gradients of each image, after centering and cropping them around the respective nucleus.
Concerning hybrid methodologies, the explicit use of both deep and conventional machine learning to detect and classify cervical cells is scarce, however, some approaches use both types of algorithms for other related tasks. For instance, Jia, Li and Zhang [14] proposed a novel system to classify cervical cells using a modified LeNet-5 CNN for feature extraction and an SVM as the final classifier, achieving better results than when using only CNN for both tasks. In a more exploratory study, Khamparia et al. [15] developed a framework for the classification of cervical cells in which several CNN and ML algorithms were tested as feature extraction mechanisms and classifiers, respectively. From the experiments performed, the combination of the ResNet50 with the random forest (RF) ensemble achieved the highest accuracy. Sarwar, Sharma and Gupta [16] also proposed a hybrid ensemble method which combined the output of 15 conventional ML and DL algorithms, consistently achieving the highest accuracy when compared to the individual use of each algorithm. Gautam et al. [17] used a different approach in which the cervical cells’ nuclei were segmented using conventional CV methods and classified in a hierarchical fashion using an AlexNet.
In a truly hybrid approach, Zhu et al. [18] proposed a system composed of a YOLOv3 network to detect the cervical lesions, followed by a more accurate reclassification of the detected targets using an Xception model. The final classification was performed according to The Bethesda System (TBS) [19] through two ensembled XGboost models, obtaining a specificity and sensitivity comparable to human cytologists. Furthermore, the system’s area under the receiver operating characteristic curve tended to increase with the number of models used, suggesting that their combination in hybrid pipelines is a step in the right direction.
3 Methodology
The proposed hybrid system, outlined in Fig. 1, follows a deep learning and a traditional machine learning approach for the detection and classification stages, respectively. Its design takes into consideration current domain knowledge by discerning between lesions with similar severity levels in the detection phase and using additional nuclei-specific features in the final classification.
3.1 Dataset
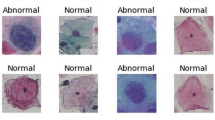
Due to the lack of public annotated data aimed at both detecting and classifying cervical lesions with the intended detail level, this work was developed using a private dataset comprehending images of microscopic fields of view of LBC samples with a resolution of 1920 \(\times \) 2560 pixels, acquired with a \(\mu \)SmartScope [20] - a portable device based on a smartphone for the automatic acquisition of microscopic images. These were obtained from 21 samples and annotated by a specialist from Hospital Fernando Fonseca (HFF). The annotations are bounding boxes surrounding the regions of interest (individual or clumps of squamous cells) and the respective class according to TBS [19]: atypical squamous cells of undetermined significance (ASC-US), low-grade squamous intraepithelial lesion (LSIL), atypical squamous cells, cannot rule out high-grade squamous intraepithelial lesion (ASC-H), high-grade squamous intraepithelial lesion (HSIL) and squamous cell carcinoma (SCC). A few examples of these classes can be seen in Fig. 2 and their distribution in Table 1, for both the entire dataset and the training set (after balancing operations).
Data Preparation. The samples are first divided, based on their diagnosis outcome, into an overall training and a test set (80% and 20%, respectively) to guarantee that images from the same patient are kept in the same set. The overall training set is further divided into several training and validation subsets by applying a stratified 5-fold cross-validation procedure to the images of each sample. The balancing of this subset is achieved through data augmentation techniques, more specifically rotations of 90, 180 and 270\(^\circ \), vertical and horizontal flipping, and blurring and sharpening of the patches containing the under-represented classes. Despite this, considering the extremely low number of examples of the SCC class, we opted to merge it with the HSIL class due to their similarity. In addition to these operations, a random down-sampling is applied on the empty patches, i.e., patches without annotations, to ensure the inclusion for each image of at least one empty patch.
3.2 Experimental Pipeline
The system’s input consists of separate images which undergo a pre-processing stage where they are cropped around the optic disk and split into multiple 320 \(\times \) 320 pixels non-overlapping patches. Then, to minimize the number of ROIs that are divided between patches, the training and validation patches are adjusted: depending on the ROI’s size, either the patch is enlarged to capture it entirely, or the fraction of the ROI is only considered if its area is equal to 10% or more of the entire ROI.
Afterwards, the detection of the abnormal cervical lesions is performed through a RetinaNet with a ResNet50 backbone pre-trained on the COCO dataset [21]. In order to optimize the network’s performance, a random hyper-parameter search process was conducted in which several settings were tuned, namely the minimum score threshold, maximum number of detections, warm-up and base learning rate, and batch size. However, due to the long training times of the neural networks, only three iterations of the 5-fold cross-validation procedure are selected in order to enable the execution of more experiments. As for the hyper-parameters that define the anchor boxes used to generate object candidates, these were modified based on the k-means clustering of the ground-truth bounding boxes’ scales and aspect ratios for the overall training set. A maximum intersection over union (IoU) threshold of 0.4 was also set since abnormal regions do not have a high degree of overlap. These experiments were performed in a Linux Server featuring one NVIDIA T4 GPU with 16 GB of RAM over approximately 300 epochs and made use of the Adam optimizer together with the smooth L1 loss for the regions’ localization, and the focal cross-entropy loss for their classification in two groups: low-grade lesions, composed of the LSIL and ASC-US classes, and high-grade lesions, composed of the HSIL-SCC and ASC-H classes. This first classification takes into account the direct dependence between the class composition and the severity level of the labels in each group, embedding clinical knowledge in the development of the pipeline.
The nuclei in these detected regions are segmented through an iterative thresholding algorithm based on the work of Phoulady et al. [5] which comprises the application of global thresholds of increasing value to the image in an iterative manner, accepting or discarding new regions based on their solidity and bounding box area. In this context, the solidity of a region refers to the measure of its concavity and is calculated by dividing its area by the area of its convex hull.
From the region and the segmented nuclei, a total of 17 geometrical, 10 colour and 34 texture features are extracted, partially inspired by the work of [22] and also due to their relevance to the clinical diagnosis. The geometrical features are composed of specific properties of the nuclei, more specifically their segmented, bounding box and convex hull areas, bounding box aspect ratio, maximum and minimum diameters, equivalent diameter, solidity, extent, minimum enclosing circle area ratio, elliptical symmetry, compactness index, principal axis ratio, eccentricity and irregularity index, the latter characterized by the difference between the maximum and minimum diameters. The colour features are extracted from the CIELCh colour space and are composed of the energy, mean, standard deviation, skewness and range of the chroma and hue channels. Lastly, the texture features are based on the dissimilarity, homogeneity, energy and correlation of the grey level co-occurrence matrices.
These features are then fed to two classifiers, one for each abnormality level detected in the previous phase. Two model types were tested for this stage: SVM and random forest. Both types underwent a process of hyper-parameter optimization, namely a grid search for the SVM and a random search for the RF (the difference between the methods stems from the longer training times of the RF models, making a grid search unfeasible). To assess the influence of the properties of the nuclei structures in the developed models, two types of feature sets were used in the experiments: one with only features extracted from the detected region as a whole - the baseline set -, and another with added nuclei features. These experiments were also repeated for the subset of the regions containing at least one nucleus to investigate if the cases in which these were absent from the region significantly impacted the performance of the algorithms. All experiments were performed using a 5-fold cross-validation procedure. It is also worth noting that the features used by the random forest algorithms remained unchanged while the ones provided to the SVM were normalized since this algorithm is based around the distance between the data points from different classes, thus not being scale-invariant.
Finally, the output of this stage (and the pipeline) is the label not only of the detected regions but of the respective patches, source images and samples as well. The diagnosis outcome of a sample is determined as the most severe lesion level found in the objects detected in all the patches extracted from the multiple source images of that sample.
4 Results
This section provides a performance evaluation of the experiments previously detailed for the lesion detection and classification modules individually and the complete system.
4.1 Lesion Detection
The system’s performance for the detection task was assessed through the COCO competition metrics [21]. From the hyper-parameter combinations tested at this stage, the best-performing one obtained a mean average precision (mAP@0.5 IoU) of 0.15 and an average recall (AR@10) of 0.42, with a base and warm-up learning rates of 0.00003 and 0.00001, correspondingly, and a minimum score, maximum detections and batch size of 0, 10 and 16, respectively, during the adapted 5-fold cross-validation. The analysis of the detections performed by RetinaNet (a couple of which are present in Figs. 3a and 3b) provides some insights regarding how certain dataset properties influenced the apparent overall low performance obtained. In particular, the high number of false positives, even in situations where the falsely detected region presented characteristics similar to the ones found in abnormal regions, may be due to the fact that abnormal regions that do not belong to a clear lesion level are not annotated in the dataset, as the specialists cannot accurately propose a classification for them.
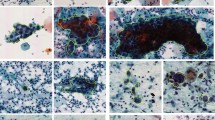
Another possible cause is the presence of squamous cells of different types that possess similar abnormality characteristics, as illustrated in Fig. 4, making them difficult to differentiate. This difficulty extends to the annotation process as well, further increasing its subjectivity. Moreover, there can be a substantial morphological variety between cells of the same type (Fig. 5), hindering the learning process by the network, especially when considering the limited amount of available data.
The high number of false negatives is also related to the presence of uninformative fractions of bounding boxes, such as the one displayed in Fig. 3c. These can be generated during the partitioning of the image into patches since our method does not take into account the image’s characteristics. This is especially noticeable in the test set, where the patches are not pre-adjusted as in the overall training set, leading to an increased number of uninformative regions and consequently to a lower mAP@0.5 IoU of 1% and an AR@10 of 27%.
4.2 Lesion Classification
The results of the classification module were first calculated for each class (pertinent to each type of classifier and at the ROI level) and then averaged together. These are displayed in Table 2 for each feature set and classifier.
It is possible to observe that the use of nuclei features had a greater influence on the low-grade regions than on the high-grade ones, where the performance of the SVM classifiers using the entire feature set was even lower than the models using only the region features. Consequently, the best-performing classifiers for each lesion level used different sets of features, with the low-grade model (RF) using both region and nuclei features, while the high-grade model (SVM) only made use of the former. These variations can be linked to the intrinsic morphological differences between the regions of each type. Although not presented in detail here, this pattern was also noticed in the experiments with only the nuclei-possessing regions, indicating that it was not derived from the absence of nuclei in certain ones.
As such, the models that obtained the highest cross-validation performance were used as the system’s final classifiers, with the low and high-grade classifiers obtaining an accuracy, precision, recall and F1 score of 0.79, 0.56, 0.50 and 0.46, and of 0.53, 0.75, 0.55 and 0.42, respectively, on the test set. Much like in the previous detection stage, the decreased performance here is also partially related to the presence of uninformative ground truth bounding boxes. Still, the possibility of overfitting having occurred cannot be ruled out due to the considerable performance difference between the cross-validation and test sets.
From Table 3 it is possible to observe that the proposed system’s classifiers are still distant from the best performing methods accuracy-wise even though it achieved similar results to the hybrid approach proposed by Sarwar, Sharma and Gupta [16] while also outperforming the most realistic configuration by Prum, Handayani and Boursier [13]. However, it is difficult to directly compare our work to the others due to the use of different datasets.
4.3 Complete System
The complete pipeline was evaluated at four different levels: regions of interest, patches, source images and samples. This is accomplished by assigning the most severe label detected in a given level to the level immediately above in the same image hierarchy. The reasoning behind this method is to try to mimic (to some extent) the evaluation process performed by the specialists since their final diagnosis relies on the analysis of multiple fields of view and specific regions within them for each patient. With this in mind, the results obtained on the test set are presented in Table 4 for each level.
As evidenced by the results, the general performance improves from the region level up to the source image evaluation (except for a slight decrease in F1 from the patch to the source image level), as the individual errors pertaining to the ROIs cease to have such a substantial impact. However, there is a performance decrease from the source image to the sample level. The analysis of the final, sample-wise outputs of the system (Table 5) reveals that there is a clear tendency to assign samples with severity labels higher than their respective ground truth. This is linked to the method described above in which a level’s classifications are passed up in the image hierarchy since it only takes one example with a severity higher than the other ones for the following level to be classified with that more severe label. Even though this method is similar to the one used by the human specialists, this drawback is not as evident in that case since they usually need multiple instances of a class (or a clear example of one) before attributing a final diagnosis.
Furthermore, over half of the samples were misclassified or falsely signalled as abnormal in the detection stage. As the exclusion of the examples incorrectly classified by the detection model enables a substantial performance increase of the system (Table 4), it becomes clear that the main performance bottleneck occurs during the detection of the abnormal regions. Even though there can be multiple reasons for this, as in the classification stage, the most limiting factor is probably the dataset itself. The relatively low number of original examples, allied to the subjective annotation process, make this a challenging task, hindering not only the training of the algorithms but their proper evaluation as well, further reinforcing the need for innovative strategies, such as hybrid approaches, to overcome these challenges.
Nonetheless, despite the system’s limitations (such as the low mAP obtained during the detection phase) that prevent its deployment as a fully automated tool, the higher recall levels achieved in the cross-validation demonstrate its potential for usage as a tool to support the analysis of the medical specialists, possibly increasing the sensitivity of their process.
5 Conclusions
In this work, a hybrid system for the examination of cervical cytology images was proposed as a way of overcoming the limitations of a private dataset and integrating clinical knowledge in the pipeline design. A RetinaNet model was used for the detection of abnormal regions and their separation in low and high-grade lesions, with random forest and SVM classifiers being applied to each of the detected regions for the specification of the final lesion class. These classification algorithms were trained using geometrical, colour and texture features extracted from each region and from the nuclei within them, which were previously segmented through an iterative thresholding algorithm.
The detection and classification modules were first evaluated separately, followed by the assessment of the complete system. While the detection module only achieved a mAP@0.5 IoU of 0.15 and an AR@10 of 0.42, the classification models were able to attain F1 scores of 0.75 and 0.79 for the low and high-grade lesions, respectively, during the 5-fold cross-validation. Moreover, the use of nuclei features during this stage resulted in a performance boost of the low-grade lesions’ classifier. Despite this, both modules’ performance dropped significantly in the test set, likely due to the substantial presence of uninformative bounding boxes derived from the partitioning of each image into patches, indicating a need for additional constraints to accept or discard fragments of bounding boxes, such as a textural analysis of the region.
As for the performance of the complete system, it generally increases with the hierarchy level. However, a noticeable decrease occurs from the source image to the sample level due to misclassifications with high severity labels occurring in the previous levels. As such, alternative strategies to aggregate the results of previous levels should be explored, potentially leading to a more reliable system.
An analysis of the system’s outputs also revealed that a significant portion of the misclassifications resulted from the erroneous identification of the lesion level by the detection module, indicating that this represents the main performance bottleneck. In this manner, hybrid approaches, such as the one proposed, reveal increased potential to tackle challenges of this nature, especially in environments where the drawbacks of the DL-only methods are not so easily overcome. Additionally, despite its limitations, the proposed system demonstrated that it can still prove helpful as a support tool for the inspection of cervical samples by medical specialists, potentially increasing the sensitivity of their analysis process.
References
World Health Organization. Cervical Cancer. https://www.who.int/health-topics/cervical-cancer. Accessed 29 June 2021
Bengtsson, E., Malm, P.: Screening for cervical cancer using automated analysis of pap-smears. Computational and mathematical methods in medicine 2014 (2014)
Conceição, T., Braga, C., Rosado, L., Vasconcelos, M.J.M.: A review of computational methods for cervical cells segmentation and abnormality classification. Int. J. Mol. Sci. 20(20), 5114 (2019)
Ushizima, D.M., Bianchi, A.G., Carneiro, C.M.: Segmentation of subcellular compartments combining superpixel representation with voronoi diagrams. Technical report, Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States) (2015)
Phoulady, H.A., Goldgof, D.B., Hall, L.O., Mouton, P.R.: An approach for overlapping cell segmentation in multi-layer cervical cell volumes. The second overlapping cervical cytology image segmentation challenge-IEEE ISBI (2015)
Kuko, M., Pourhomayoun, M.: Single and clustered cervical cell classification with ensemble and deep learning methods. Inf. Syst. Front. 22(5), 1039–1051 (2020)
Du, J., Li, X., Li, Q.: Detection and classification of cervical exfoliated cells based on faster R-CNN. In: 2019 IEEE 11th International Conference on Advanced Infocomm Technology, ICAIT 2019, pp. 52–57. Institute of Electrical and Electronics Engineers Inc., October 2019
Zhou, M., Zhang, L., Du, X., Ouyang, X., Zhang, X., Shen, Q., Wang, Q.: Hierarchical and Robust Pathology Image Reading for High-Throughput Cervical Abnormality Screening. In: Liu, M., Yan, P., Lian, C., Cao, X. (eds.) MLMI 2020. LNCS, vol. 12436, pp. 414–422. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59861-7_42
Zhang, L., Lu, L., Nogues, I., Summers, R.M., Liu, S., Yao, J.: DeepPap: deep convolutional networks for cervical cell classification. IEEE J. Biomed. Health Inf. 21(6), 1633–1643 (2017)
Kwon, M., Kuko, M., Pourhomayoun, M., Martin, V., Kim, T., Martin, S.: Multi-label Classification of Single and Clustered Cervical Cells Using Deep Convolutional Networks. California State University, Los Angeles (2018)
Ghoneim, A., Muhammad, G., Hossain, M.S.: Cervical cancer classification using convolutional neural networks and extreme learning machines. Futur. Gener. Comput. Syst. 102, 643–649 (2020)
Su, J., Xu, X., He, Y., Song, J.: Automatic detection of cervical cancer cells by a two-level cascade classification system. Analytical Cellular Pathology 2016 (2016)
Sophea, P., Handayani, D.O.D., Boursier, P.: Abnormal cervical cell detection using hog descriptor and SVM classifier. In: 2018 Fourth International Conference on Advances in Computing, Communication & Automation (ICACCA), pp. 1–6. IEEE (2018)
Jia, A.D., Li, B.Z., Zhang, C.C.: Detection of cervical cancer cells based on strong feature CNN-SVM network. Neurocomputing 411, 112–127 (2020)
Khamparia, A., Gupta, D., de Albuquerque, V.H.C., Sangaiah, A.K., Jhaveri, R.H.: Internet of health things-driven deep learning system for detection and classification of cervical cells using transfer learning. The Journal of Supercomputing 76(11), 8590–8608 (2020). https://doi.org/10.1007/s11227-020-03159-4
Sarwar, A., Sharma, V., Gupta, R.: Hybrid ensemble learning technique for screening of cervical cancer using papanicolaou smear image analysis. Personalized Med. Universe 4, 54–62 (2015)
Gautam, S., Jith, N., Sao, A.K., Bhavsar, A., Natarajan, A., et al.: Considerations for a pap smear image analysis system with CNN features. arXiv preprint arXiv:1806.09025 (2018)
Zhu, X., et al.: Hybrid AI-assistive diagnostic model permits rapid TBS classification of cervical liquid-based thin-layer cell smears. Nat. Commun. 12(1), 1–12 (2021)
Birdsong, G.G., Davey, D.D.: The bethesda system for reporting cervical cytology. In: The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes, pp. 1–28. Springer International Publishing, January 2015
Rosado, L., et al.: Smartscope: towards a fully automated 3d-printed smartphone microscope with motorized stage. In: International Joint Conference on Biomedical Engineering Systems and Technologies, pp. 19–44. Springer (2017)
COCO - Common Objects in Context. https://cocodataset.org/. Accessed 16 May 2021
Rosado, L., Da Costa, J.M.C., Elias, D., Cardoso, J.S.: Mobile-based analysis of malaria-infected thin blood smears: automated species and life cycle stage determination. Sensors 17(10), 2167 (2017)
Acknowledgments
This work was done under the scope of project “CLARE: Computer-Aided Cervical Cancer Screening” and financially supported by FEDER through Operational Competitiveness Program – COMPETE 2020 and by National Funds through Foundation for Science and Technology FCT/MCTES (POCI-01-0145-FEDER-028857). The authors would also like to thank Hospital Fernando Fonseca’s and Portuguese Oncology Institute’s Pathology Anatomy Services.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this paper
Cite this paper
Silva, E.L., Sampaio, A.F., Teixeira, L.F., Vasconcelos, M.J.M. (2021). Cervical Cancer Detection and Classification in Cytology Images Using a Hybrid Approach. In: Bebis, G., et al. Advances in Visual Computing. ISVC 2021. Lecture Notes in Computer Science(), vol 13018. Springer, Cham. https://doi.org/10.1007/978-3-030-90436-4_24
Download citation
DOI: https://doi.org/10.1007/978-3-030-90436-4_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-90435-7
Online ISBN: 978-3-030-90436-4
eBook Packages: Computer ScienceComputer Science (R0)