Abstract
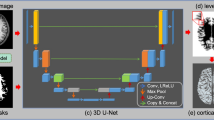
Commonly-used tools for cortical reconstruction and parcellation, such as FreeSurfer, are central to brain surface analysis but require extensive computation times. This paper proposes SegRecon, a fast learning approach where an integrated end-to-end deep learning method does simultaneously reconstruct and segment cortical surfaces directly from an MRI volume, all in a single step. We train a volume-based neural network to predict, for each voxel, the signed distance to the white-to-grey-matter interface along with its corresponding spherical representation in the registered atlas space. The continuous representation of the spherical coordinates enables our approach to naturally extract an implicit isolevel surface for its reconstruction and obtain the parcel labels from the spherical atlas. We illustrate the advantages of our method with thorough experiments on the MindBoggle dataset. Our parcellation results show more than 4% improvements in average Dice accuracy with respect to FreeSurfer and a drastic speed-up from hours to seconds of computation.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bazin, P.L., Pham, D.L.: Topology correction of segmented medical images using a fast marching algorithm. Comput. Meth. Prog. Biomed. 88(2), 182–190 (2007)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: MICCAI (2016)
Cruz, R.S., Lebrat, L., Bourgeat, P., Fookes, C., Fripp, J., Salvado, O.: DeepCSR: a 3D deep learning approach for cortical surface reconstruction. arXiv preprint arXiv:2010.11423 (2020)
Dahnke, R., Yotter, R.A., Gaser, C.: Cortical thickness and central surface estimation. Neuroimage 65, 336–348 (2013)
Desikan, R.S., et al.: An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31(3), 968–980 (2006)
Fischl, B., et al.: Automatically parcellating the cortex. Cereb. Cortex 14(1), 11–22 (2004)
Fischl, B., Sereno, M.I., Dale, A.M.: Cortical surface-based analysis: Ii: inflation, flattening, and a surface-based coordinate system. Neuroimage 9(2), 195–207 (1999)
Glasser, M.F., et al.: A multi-modal parcellation of human cerebral cortex. Nature 536(7615), 171–178 (2016)
Gopinath, K., Desrosiers, C., Lombaert, H.: Graph convolutions on spectral embeddings for cortical surface parcellation. Med. Image Anal. 54, 297–305 (2019)
Gopinath, K., Desrosiers, C., Lombaert, H.: Graph domain adaptation for alignment-invariant brain surface segmentation. arXiv preprint arXiv:2004.00074 (2020)
He, R., Gopinath, K., Desrosiers, C., Lombaert, H.: Spectral graph transformer networks for brain surface parcellation. In: ISBI (2020)
Henschel, L., Conjeti, S., Estrada, S., Diers, K., Fischl, B., Reuter, M.: Fastsurfer-a fast and accurate deep learning based neuroimaging pipeline. NeuroImage (2020)
Kim, J.S., et al.: Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. Neuroimage 27(1), 210–221 (2005)
Kingma, D.P., Ba, J.: Adam: stochastic optimization. In: ICLR (2014)
Klein, A., et al.: Mindboggling morphometry of human brains. PLOS Comput. Biol. 13(2), e1005350 (2017)
Klein, A., Tourville, J.: 101 labeled brain images and a consistent human cortical labeling protocol. Front. Neurosci. 6, 171 (2012)
Kriegeskorte, N., Goebel, R.: An efficient algorithm for topologically correct segmentation of the cortical sheet in anatomical MR volumes. NeuroImage 14(2), 329–346 (2001)
Lombaert, H., Criminisi, A., Ayache, N.: Spectral forests: learning of surface data, application to cortical parcellation. In: MICCAI (2015)
López-López, N., Vázquez, A., Poupon, C., Mangin, J.F., Ladra, S., Guevara, P.: GeoSP: a parallel method for a cortical surface parcellation based on geodesic distance. In: EMBC (2020)
Lorensen, W.E., Cline, H.E.: Marching cubes: a high resolution 3D surface construction algorithm. ACM SIGGRAPH Comput. Graph. 21(4), 163–169 (1987)
Park, J.J., Florence, P., Straub, J., Newcombe, R., Lovegrove, S.: DeepSDF: Learning continuous signed distance functions for shape representation. In: CVPR (2019)
Perona, P., Malik, J.: Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 12(7), 629–639 (1990)
Querbes, O., et al.: Early diagnosis of Alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain 132(Pt 8), 2036–2047 (2009)
Shattuck, D.W., Leahy, R.M.: Brainsuite: an automated cortical surface identification tool. Med. Image Anal. 6(2), 129–142 (2002)
Wu, Z., et al.: Intrinsic patch-based cortical anatomical parcellation using graph convolutional neural network on surface manifold. In: MICCAI (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Gopinath, K., Desrosiers, C., Lombaert, H. (2021). SegRecon: Learning Joint Brain Surface Reconstruction and Segmentation from Images. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12907. Springer, Cham. https://doi.org/10.1007/978-3-030-87234-2_61
Download citation
DOI: https://doi.org/10.1007/978-3-030-87234-2_61
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87233-5
Online ISBN: 978-3-030-87234-2
eBook Packages: Computer ScienceComputer Science (R0)