Abstract
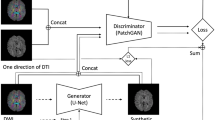
Unpaired image-to-image translation has been applied successfully to natural images but has received very little attention for manifold-valued data such as in diffusion tensor imaging (DTI). The non-Euclidean nature of DTI prevents current generative adversarial networks (GANs) from generating plausible images and has mainly limited their application to diffusion MRI scalar maps, such as fractional anisotropy (FA) or mean diffusivity (MD). Even if these scalar maps are clinically useful, they mostly ignore fiber orientations and therefore have limited applications for analyzing brain fibers. Here, we propose a manifold-aware CycleGAN that learns the generation of high-resolution DTI from unpaired T1w images. We formulate the objective as a Wasserstein distance minimization problem of data distributions on a Riemannian manifold of symmetric positive definite 3 \(\times \) 3 matrices SPD(3), using adversarial and cycle-consistency losses. To ensure that the generated diffusion tensors lie on the SPD(3) manifold, we exploit the theoretical properties of the exponential and logarithm maps of the Log-Euclidean metric. We demonstrate that, unlike standard GANs, our method is able to generate realistic high-resolution DTI that can be used to compute diffusion-based metrics and potentially run fiber tractography algorithms. To evaluate our model’s performance, we compute the cosine similarity between the generated tensors principal orientation and their ground-truth orientation, the mean squared error (MSE) of their derived FA values and the Log-Euclidean distance between the tensors. We demonstrate that our method produces 2.5 times better FA MSE than a standard CycleGAN and up to 30% better cosine similarity than a manifold-aware Wasserstein GAN while synthesizing sharp high-resolution DTI.
This work was supported financially by the Réseau de Bio-Imagerie du Québec (RBIQ), the Research Council of Canada (NSERC), the Fonds de Recherche du Québec (FQRNT), ETS Montreal, and NVIDIA with the donation of a GPU.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Arjovsky, M., Chintala, S., Bottou, L.: Wasserstein GAN (2017)
Arsigny, V., Fillard, P., Pennec, X., Ayache, N.: Log-Euclidean metrics for fast and simple calculus on diffusion tensors. Mag. Reson. Med. (2006)
Glasser, M.F., Sotiropoulos, S.N., Wilson, J.A., Coalson, T.S., Fischl, B., Andersson, J.L., Xu, J., Jbabdi, S., Webster, M., Polimeni, J.R., Van Essen, D.C., Jenkinson, M.: The minimal preprocessing pipelines for the human connectome project. NeuroImage (2013)
Goodfellow, I.J., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., Courville, A., Bengio, Y.: Generative adversarial nets. In: Advances in Neural Information Processing Systems. Neural Information Processing Systems Foundation (2014)
Gu, X., Knutsson, H., Nilsson, M., Eklund, A.: Generating Diffusion MRI Scalar Maps from T1 Weighted Images Using Generative Adversarial Networks. Technical Report (2019)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society (2016)
Huang, Z., Van Gool, L.: A riemannian network for SPD matrix learning. 31st AAAI Conference on Artificial Intelligence, AAAI 2017 (2017)
Huang, Z., Wu, J., Van Gool, L.: Manifold-valued image generation with wasserstein generative adversarial nets. In: Proceedings of the AAAI Conference on Artificial Intelligence, pp. 3886–3893 (2019)
Ionescu, C., Vantzos, O., Sminchisescu, C.: Matrix backpropagation for deep networks with structured layers. Technical Report (2015)
Jenkinson, M., Bannister, P., Brady, M., Smith, S.: Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage (2002)
Jenkinson, M., Beckmann, C.F., Behrens, T.E.J., Woolrich, M.W., Smith, S.M.: Review FSL. NeuroImage (2012)
Jenkinson, M., Smith, S.: A global optimisation method for robust affine registration of brain images. Med. Image Anal. (2001)
Jiang, H., Van Zijl, P.C.M., Kim, J., Pearlson, G.D., Mori, S.: DtiStudio: Resource program for diffusion tensor computation and fiber bundle tracking (2005)
Kingma, D.P., Ba, J.L.: Adam: A method for stochastic optimization. In: 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings. International Conference on Learning Representations, ICLR (2015)
Pfefferbaum, A., Sullivan, E.V.: Increased brain white matter diffusivity in normal adult aging: relationship to anisotropy and partial voluming. Mag. Reson. Med. Official J. Int. Soc. Mag. Reson. Med. (2003)
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) (2015)
Schyboll, F., Jaekel, U., Weber, B., Neeb, H.: The impact of fibre orientation on t1-relaxation and apparent tissue water content in white matter. Mag. Reson. Mater. Phys. Biol. Med. 31(4), 501–510 (2018)
Sotiropoulos, S.N., Jbabdi, S., Xu, J., Andersson, J.L., Moeller, S., Auerbach, E.J., Glasser, M.F., Hernandez, M., Sapiro, G., Jenkinson, M., Feinberg, D.A., Yacoub, E., Lenglet, C., Van Essen, D.C., Ugurbil, K., Behrens, T.E.: Advances in diffusion MRI acquisition and processing in the human connectome project. NeuroImage (2013)
Taoka, T., Morikawa, M., Akashi, T., Miyasaka, T., Nakagawa, H., Kiuchi, K., Kishimoto, T., Kichikawa, K.: Fractional anisotropy-threshold dependence in tract-based diffusion tensor analysis: evaluation of the uncinate fasciculus in alzheimer disease. Am. J. Neuroradiol. 30(9), 1700–1703 (2009)
Van Essen, D.C., Smith, S.M., Barch, D.M., Behrens, T.E., Yacoub, E., Ugurbil, K.: The WU-minn human connectome project: an overview. NeuroImage (2013)
Yi, X., Walia, E., Babyn, P.: Generative adversarial network in medical imaging: a review. Med. Image Anal. 58 (2019)
Zhong, J., Wang, Y., Li, J., Xue, X., Liu, S., Wang, M., Gao, X., Wang, Q., Yang, J., Li, X.: Inter-site harmonization based on dual generative adversarial networks for diffusion tensor imaging: application to neonatal white matter development. BioMed. Eng. Online 19 (2020)
Zhu, J.Y., Park, T., Isola, P., Efros, A.A.: Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Anctil-Robitaille, B., Desrosiers, C., Lombaert, H. (2021). Manifold-Aware CycleGAN for High-Resolution Structural-to-DTI Synthesis. In: Gyori, N., Hutter, J., Nath, V., Palombo, M., Pizzolato, M., Zhang, F. (eds) Computational Diffusion MRI. Mathematics and Visualization. Springer, Cham. https://doi.org/10.1007/978-3-030-73018-5_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-73018-5_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-73017-8
Online ISBN: 978-3-030-73018-5
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)