Abstract
In arteriovenous malformation (AVM) surgery, vessel structures should be well evaluated with angiography. However, with conventional angiography, it is sometimes difficult to distinguish each feeder and its feeding territory in the nidus. In this study, we used two software systems to create three-dimensional (3D) fusion images using multiple imaging modalities and evaluated their clinical use. In the AVM patient, data were obtained from 3D rotational angiography, rotational venography, computed tomography (CT), and magnetic resonance imaging (MRI) and superimposed into 3D fusion images using imaging software (iPLAN and Avizo). Virtual surgical fields that were quite similar to the real ones were also created with these software programs. Compared with fusion images by iPLAN, those by Avizo have higher resolution and can demarcate not only each feeder but also its supplying territory in the nidus with different colors.
In conclusion, 3D fusion images in AVM surgery are helpful for simulation, even though it takes time and requires special skill to create them.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
Introduction
In AVM surgery, evaluation of angiostructure is essential to make a good surgical strategy. With conventional two-dimensional digital subtraction angiography (DSA), it is sometimes difficult to understand AVM structure stereoscopically. Three-dimensional rotational angiography (3D-RA) gives us much more precise information about the relationships of each feeder, drainer, and nidus, but when the AVM is supplied by a multiple vascular territory (e.g., middle cerebral artery and posterior cerebral artery), all the details cannot be seen in one image. For the surgical simulation, the location of the nidus in the brain is also important so as to decide the surgical approach. Recent image fusion technologies have helped surgeons create virtual surgical fields, especially in brain tumors [1,2,3]. In this article the authors introduced 3D fusion images in AVM surgeries and evaluated their clinical use.
Methods
Image data, including 3D-RA, 3D-rotational venography (RV), comported tomography (CT), and magnetic resonance imaging (MRI), was obtained from AVM patients. Three-dimensional-RA and RV were performed with a C-arm angiography unit (Allura XperFD 20/10; Philips Medical Systems, Best, the Netherlands). The C-arm rotated through 240° at 55°/s and obtained 120 images on a 17-in. FOV during contrast injection. MRI was performed with a 3.0-T system for the head (Ingenia 3.0T; Philips Healthcare, Andover, MA.). Fluid-attenuated inversion recovery (FLAIR) in MRI was acquired with an eight-channel head coil. CT was done with a 64-section CT scanner (Aquilion; Toshiba Medical Systems, Tokyo, Japan). The slice thickness was 1 mm.
Image data was coded in digital imaging and communication in medicine (DICOM) format and was imported to the two different imaging applications. In this study, we used iPLAN cranial (BrainLab, Germany) and Avizo (Visualization Science Group, Bordeaux, France) for 3D image reconstruction using a previously reported method [1, 2]. In iPLAN, the 3D model was constructed with a volume rendering method with autosegmentation, whereas Avizo used a hybrid method combining surface- and volume-rendering methods, and manual segmentation was used to distinguish the small anatomical structures, such as the feeding arteries, from surrounding noise.
Results
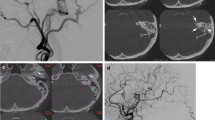
The 3D fusion images of the right parietal lobe AVM for a representative patient are illustrated in Fig. 1. The fusion image with iPLAN has lower resolution and the feeders, nidus, and drainers are not distinguishable because these three vasculatures are visualized in the same time phase in case of arterio-venous shunting disease. Avizo created higher resolution images, and feeders, drainers, and normal vessels are visualized clearly with different colors. Also, each feeder and its supplying territory in the nidus are distinguished, and surgical simulation becomes possible after adding the MRI (FLAIR) and CT images (Fig. 2). On the other hand, the time required for creation is much shorter in iPLAN (approximately 1 h) than Avizo (more than 3 h).
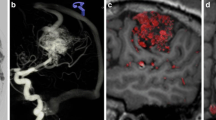
Three-dimensional fusion images of the same patient by Avizo and comparison to the surgical field. (a, b) Not only each feeder but also its supplying territory in the nidus is well demarcated. Feeders from anterior cerebral arteries are colored red and cyan, those from middle cerebral arteries are green, and those from posterior cerebral arteries are pink. (c, d) Surgical simulation using fusion images. The feeder existing just behind the main drainer (arrows) is well recognized. (e) Real surgical field in the same patient. The cortical artery feeders were quite similar to fusion image (c)
Discussion
In AVM surgery, understanding the angiostructure is essential for planning good surgical strategy. In principle, feeders should be occluded before the dissection and obliteration of the gliotic tissue around the nidus. However, in conventional angiography, and especially with high-flow AVMs, it is sometimes difficult to identify each feeder and its anatomical relationship with the nidus and drainers. Three dimensional-RA images give us much more precise information about the angiostructure of AVMs, and have become prerequisites for the surgery. However, in the real surgical field, we have to dissect the nidus from the brain, and the feeders may be found on the brain surface or in the sulci or fissures; therefore it is also necessary to evaluate the anatomical relationship between the AVM and the brain. Surgeons must currently use their experience and knowledge to mentally reconstruct 3D fusion images from different modalities for this purpose. In this study, we created two types of 3D fusion images in AVM patients with different imaging software and compared their efficacy in the surgical simulations. Images by iPLAN were easily created without much time or AVM surgery experience, because this software enables the automatic segmentation of DICOM data. The resultant 3D fusion image is highly versatile, the quality is similar between creators, and it can be introduced to the navigation system (BrainLab), all of which make surgical simulation easier for less experienced surgeons. On the other hand, the resolution is not so high and each feeder and its supplying territory in the nidus are not distinguishable. Moreover, it is difficult to find “hidden feeders” existing just behind the drainer, which we sometimes encounter in a clinical setting, because feeders and drainers are recognized in the same color. In contrast, 3D fusion images by Avizo are created by both surface and volume rendering methods, so that they have a much higher resolution and the contrast between feeders, nidus, and drainers are much better than with the images by iPLAN. Therefore, we can identify all feeders even if they are close to the drainers or “en passage” arteries (Fig. 2). Avizo can also discriminate not only each feeder but also its supplying territory in the nidus with different colors, helping us to understand which feeders impinge more on the nidus (Fig. 2). According to these data, we can then decide which feeders have priority for surgical or endovascular occlusion. Adding MRI data to the Avizo images produces a virtual surgical field and surgical simulation (positioning, craniotomy, exposure of the brain, and the dissection of the nidus) becomes easy.
AVM surgical simulation with other virtual reality technologies has also been published [4, 5]. In these articles, the authors created 3D fusion images with a virtual reality simulator (Dextroscope), but they used only MR-angiography (MRA) and MR-venography (MRV) as vessel images, so the resolution of each vessel was not as high and classification of each feeder and its supplying territory in the nidus was impossible. To our knowledge, only Avizo provides adequate detail to distinguish individual feeder territories, so it may have an advantage especially in the surgical simulation of large, complex AVMs. On the other hand, the limitation of the Avizo imaging technique is that it is time-consuming because the segmentation of DICOM data and extraction of each feeder must be done manually and requires some knowledge of AVM surgery.
Conclusions
To make the surgical simulation of AVM easier, it is important to visualize all vasculatures sterically. Recent technologies have made this level of detail possible with 3D fusion images. Our study demonstrates that the Aviso 3D fusion images may be especially useful in surgical simulations for large, complex AVMs, and adoption of this technology may contribute to safer AVM surgeries.
References
Kin T, Nakatomi H, Shojima M, Tanaka M, Ino K, Mori H, Kunimatsu A, Oyama H, Saito N (2012) A new strategic neurosurgical planning tool for brainstem cavernous malformations using interactive computer graphics with multimodal fusion images. J Neurosurg 117(1):78–88
Yoshino M, Kin T, Nakatomi H, Oyama H, Saito N (2013) Presurgical planning of feeder resection with realistic three-dimensional virtual operation field in patient with cerebellopontine angle meningioma. Acta Neurochir 155(8):1391–1399
Yoshino M, Nakatomi H, Kin T et al (2017) Usefulness of high-resolution 3D multifusion medical imaging for preoperative planning in patients with posterior fossa hemangioblastoma: technical note. J Neurosurg 127(1):139–147
Ng I, Hwang PYK, Kumar D, Lee CK, Kockro RA, Sitoh YY (2009) Surgical planning for microsurgical excision of cerebral arterio-venous malformations using virtual reality technology. Acta Neurochir 151(5):453–463
Wong GKC, Zhu XL, Ahuja AT, Poon WS (2009) Stereoscopic virtual reality simulation for microsurgical excision of cerebral arteriovenous malformation: case illustrations. Surg Neurol 72(1):69–72
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this paper
Cite this paper
Hara, T., Yoshino, M. (2021). Surgical Simulation with Three-Dimensional Fusion Images in Patients with Arteriovenous Malformation. In: Esposito, G., Regli, L., Cenzato, M., Kaku, Y., Tanaka, M., Tsukahara, T. (eds) Trends in Cerebrovascular Surgery and Interventions. Acta Neurochirurgica Supplement, vol 132. Springer, Cham. https://doi.org/10.1007/978-3-030-63453-7_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-63453-7_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-63452-0
Online ISBN: 978-3-030-63453-7
eBook Packages: MedicineMedicine (R0)