Abstract
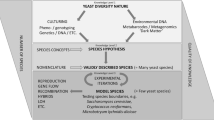
Species constitute the fundamental units of taxonomy and an ideal species definition would embody groups of genetically cohesive organisms reflecting their shared history, traits, and ecology. In contrast to animals and plants, where genetic cohesion can essentially be characterized by sexual compatibility and population structure, building a biologically relevant species definition remains a challenging endeavor in prokaryotes. Indeed, the structure, ecology, and dynamics of microbial populations are still largely enigmatic, and many aspects of prokaryotic genomics deviate from sexual organisms. In this chapter, I present the main concepts and operational definitions commonly used to designate microbial species. I further emphasize how these different concepts accommodate the idiosyncrasies of prokaryotic genomics, in particular, the existence of a core- and a pangenome. Although prokaryote genomics is undoubtedly different from animals and plants, there is growing evidence that gene flow—similar to sexual reproduction—plays a significant role in shaping the genomic cohesiveness of microbial populations, suggesting that, to some extent, a species definition based on the Biological Species Concept is applicable to prokaryotes. Building a satisfying species definition remains to be accomplished, but the integration of genomic data, ecology, and bioinformatics tools has expanded our comprehension of prokaryotic populations and their dynamics.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 The Bacterial Species Challenge
Are There Bacterial Species?
The taxonomy of microorganisms has been delayed relative to macroscopic organisms, due in part to technical reasons. Evolutionary biologists and population geneticists have originally focused their works on animals and plants, which typically engage in sexual reproduction. For these organisms, speciation mechanisms involve—directly or indirectly—the sustained interruption of gene flow between populations (Dobzhansky 1935; Mayr 1942). The maintenance of gene flow warrants the genetic cohesion of populations, but because prokaryotes do not engage in sexual reproduction stricto sensu, the definition of species has been more elusive in bacteria. It has even been suggested that bacteria cannot and need not be organized into species, but rather represent a series of organisms with different levels of divergence to one another reflecting their past history (Doolittle and Zhaxybayeva 2009; Bapteste et al. 2009). In other words, this view suggests that imposing a grouping of bacteria into species would be purely arbitrary and unreflective of any biologically-relevant process (e.g., cessation of gene flow). However, in practice, microbiologists can usually recognize and designate bacterial isolates based on their different phenotypic characteristics, and comparisons of bacterial genomes indicate that bacteria form clear clusters of highly related individuals, instead of showing a scattered distribution (Riley and Lizotte-Waniewski 2009; Caro-Quintero and Konstantinidis 2012; Konstantinidis et al. 2017), suggesting that they can be organized into species. Ecologically, bacteria can also be identified and clustered based on shared niches and properties (Shapiro and Polz 2014). Altogether, these observations indicate that bacteria can clearly be grouped into genetically and ecologically cohesive entities characteristic of “species”, although such species might not be defined based on the same criteria as for sexual organisms. The bacterial species challenge aims to determine the processes that are shaping and maintaining these clusters of cohesive entities.
Bacterial Genomics and the Case of Escherichia coli
Before the advent of genotyping methods, microbiologists had to rely exclusively on phenotypic traits to characterize and classify bacteria. Such phenotypic observations offer one criterion for building a species concept, similar to the early approaches used by naturalists to classify animals and plants. However, these early observations showed that it might not be that simple. The seminal work of Oswald Avery and colleagues had strong implications in the field of biology by identifying that DNA—not proteins—was the support of heredity (Avery et al. 1944). But this experiment and previous others further demonstrated that some phenotypic traits could be transmitted horizontally from one bacterial cell to another (Griffith 1928). Although it took several decades to fully understand the extent of horizontal gene transfer in bacteria, this challenging observation contrasted with animals and plants where traits are almost exclusively inherited vertically (i.e., from parent to offspring), indicating that something about bacteria was profoundly different. The development of genetic and genomic techniques further revealed how deeply bacterial genomics differed from animals and plants: related bacteria can differ dramatically in their gene contents and what is typically considered as a bacterial species presents a set of ubiquitous and highly similar genes, the core-genome, but also a set of accessory genes (also called dispensable, flexible, or auxiliary genes) presenting a scattered distribution (Vernikos et al. 2015). The pangenome represents the total gene diversity of a given population: this comprises the total number of distinct orthologs, including core genes and accessory genes (Tettelin et al. 2005; Medini et al. 2005; Vernikos et al. 2015).
The bacteria Escherichia coli perfectly illustrates the genomic versatility of prokaryotes. E. coli contains approximatively 4400 genes for its model strain K12 MG1655 (Hayashi et al. 2006), but other strains contain up to an additional 1000 genes encoding for a variety of functions (Hayashi et al. 2001). The comparison of only 20 strains of E. coli shows that the set of genes shared by all strains—the core-genome—is composed of approximately 2000 genes, but its pangenome approaches readily 18,000 genes (Touchon et al. 2009) and the inclusion of additional strains would necessarily increase this number, as suggested by resampling analyses (Touchon et al. 2009). These numbers indicate that over 50% of the genes of a single strain of E. coli consist of accessory genes that do not contain orthologs in the majority of all other strains. Importantly, most of these accessory genes are typically restricted to a single or a small subset of strains, but are often exchanged between strains (Groisman and Ochman 1996; Gogarten et al. 2002; Touchon et al. 2009). Many strains of E. coli possess different lifestyles and ecologies broadly ranging from environmental to commensal or pathogenic and these differences can be primarily ascribed to their specific sets of accessory genes (Luo et al. 2011). For example, virulence genes represent a category of extensively studied accessory genes and they appear to be frequently exchanged during E. coli’s evolution (Groisman and Ochman 1996; Gogarten et al. 2002).
Although E. coli strains present different phenotypes and many different assemblages of accessory genes, they still form a cohesive entity since they share a large number of core genes that are highly similar between all strains of E. coli (typically >98% of sequence identity) (Bobay et al. 2013). This situation is problematic for applying phenotype-based classifications in microbiology, as emphasized by the case of Shigella. This bacterial “genus” comprises four recognized species (i.e., S. flexneri, S. boydii, S. sonnei, and S. dysenteriae), which have been grouped based on shared phenotypic properties (i.e., they are obligate pathogens) (Rolland et al. 1998; Pupo et al. 2000; Escobar-Paramo et al. 2003). However, genomic analyses showed that Shigella possesses the same core-genome as E. coli with an average of >98% of sequence identity across core genes and core-genome phylogenies revealed that Shigella do not form a monophyletic clade (Touchon et al. 2009). What unites Shigella together is the presence of shared virulence genes (Buchrieser et al. 2000; Touchon et al. 2009), their serology, and their incapacity to ferment lactose or decarboxylate lysine (Hale and Keusch 1996). In other words, Shigella constitutes a subset of E. coli’s strains with a shared phenotype conferred by the independent gain of a common set of accessory genes by horizontal gene transfer. It is now recognized that Shigella are part of the E. coli species, but its taxonomy has not been revised. This example illustrates that the pangenome and its evolutionary dynamics represent a challenge to disentangling the complex relationship between phenotypes, ecology, and genomics in bacteria and how these characteristics correlate with taxonomy.
2 Species Concepts and Operational Definitions
Pragmatic Approaches: Sequence Thresholds
One of the goals of a taxonomy is to facilitate communication in the scientific community. To satisfy the need of a coherent microbial taxonomy, pragmatic approaches have been developed in order to define species based on genetic or genomic similarities. Although this does not directly offer insight into how and why a given set of strains constitutes a species, a threshold-based method provides a convenient means to classify strains and revise taxonomy as more comparative genomic data become available. Due to the lack of a theoretical framework of these approaches, such threshold-based methods are often said to define Operational Taxonomic Units (OTUs) rather than “species” to emphasize that this is only an operational definition.
Before the rise of the genomic era, species membership was established by shared phenotypic traits and by DNA–DNA hybridization essays, which consist of comparing a newly isolated strain to a reference strain (Brenner et al. 2000) (note that other criteria such as GC content were also considered). The recommended threshold to define species membership was set at 70% of genomic hybridization to the reference strain (Brenner et al. 2000). The emergence of sequencing technologies led to the rise of related approaches. The 16S rRNA subunit has been identified as a universal gene shared by all bacteria and archaea (Woese and Fox 1977) offering the possibility to assess prokaryotic species membership with the same gene marker across all lineages. Analyses revealed that the threshold of 70% identity based on DNA–DNA hybridization assays corresponds approximately to a threshold of 97% identity when using the 16S rRNA subunit (Stackebrandt and Goebel 1994; Ludwig and Klenk 2000; Richter and Rossello-Mora 2009). The use of 16S rRNA thresholds can be applied with ease and allows for the identification of a species by sequencing a single locus. OTU-typing based on the 16S rRNA gene became even more popular with the rise of metagenomic sequencing, where the amplification and sequencing of a fragment of the 16S rRNA gene provides a direct overview of the taxonomic diversity of a given sample without the need of cultivating any of its members. A more recent approach consists of using the entire genome of a strain to calculate the Average Nucleotide Identity (ANI) across all the genes relative to a reference genome of the species (Konstantinidis and Tiedje 2005; Richter and Rossello-Mora 2009). Because protein-coding genes are not as selectively constrained as the 16S rRNA subunit, the ANI threshold used to attain species membership has been empirically defined as 95% based on correlations with 16S sequence threshold used to define species (Konstantinidis and Tiedje 2005; Richter and Rossello-Mora 2009). Considering complete genomes obviously offers a more accurate resolution of sequence divergence.
Sequence thresholds based on single loci or entire genomes present the advantage of defining all prokaryotic species under a standardized framework, but, despite their simplicity, they suffer several technical difficulties. Sequences of the 16S rRNA subunit evolve very slowly and thus sequences from related strains or species typically display little or no informative differences (Kettler et al. 2007). Moreover, multiple copies of the 16S rRNA gene are frequently found in the same genome and they sometimes exhibit different levels of divergence (Acinas et al. 2004). In several cases, the different 16S rRNA copies present in the same genome can display remarkable levels of divergence, such as Thermoanaerobacter tengcongensis, which presents 11.6% of sequence divergence between its most different 16S rRNA copies (Acinas et al. 2004). Comparing these sequences would lead to the ironic conclusion that the same bacterial isolate should be classified into two distinct species. A more common criticism against 16S rRNA thresholds is that the divergence of the 16S rRNA gene does not always accurately reflect overall genomic divergence. For instance, the marine bacterium Prochlorococcus can be classified as a single species based on 16S rRNA sequences but some strains display only 66% genome-wide identity based on ANI methods (Zhaxybayeva et al. 2009). ANI thresholds are recognized as much more reliable criteria to define species and 16S rRNA alone is of little taxonomic value when complete genome sequences are available (Richter and Rossello-Mora 2009). However, ANI-based methods also suffer inconsistencies. Sequence identity might not be constant along the entire genome (Retchless and Lawrence 2007, 2010) and the identity thresholds used to infer gene orthology can therefore affect the overall ANI value. Perhaps more importantly, ANI metrics are frequently computed against a single reference genome to assess species membership, but the choice of reference genomes is largely arbitrary and historically contingent. In other words, species borders can vary depending on which—or how many—genomes are used as a reference. Finally, using a fixed sequence threshold does not account for the different rates of genomic evolution across phyla (Hugenholtz et al. 2016), which are dictated by parameters like mutation rates, selection coefficients, and effective population sizes (Shapiro 2014) that vary across prokaryotic lineages. Other mechanisms might further lead to differential rates of evolution such as the lack of DNA repair systems (Dorer et al. 2011). Bacterial endosymbionts notoriously evolve at faster rates due to less effective selective pressures imposed by their reduced population sizes (Moran 1996; Moran et al. 2009). As a consequence, the sequence threshold constituting a species in symbiotic bacteria likely corresponds to a different time scale in free-living bacteria (Parks et al. 2018). As a result of all these issues, applying sequence thresholds to define species is convenient but does not anchor a bacterial species concept on a solid theoretical framework.
Phylogenetic Concept
Phylogenetic approaches offer another means to classify species. As for sequence thresholds, phylogenetic methods are also a pragmatic approach to define species, although phylogenetic species are defined in the context of evolutionary history (De Queiroz and Gauthier 1994). Besides taking sequence divergence into account, phylogenies typically require species and other taxa to constitute monophyletic groups. Although the concept of monophyly is usually a key feature researched by phylogenetic approaches, it has been argued that exclusivity might be preferable over monophyly (Velasco 2009; Wright and Baum 2018). Exclusivity is defined as groups of strains/taxa that are more related to one another than other groups without being necessarily monophyletic (Velasco 2009; Wright and Baum 2018). A recent study focusing on Streptomycetaceae and Bacillus found that exclusive clades can be defined for these taxa, although no objective threshold appears universal (Wright and Baum 2018). An additional and nontrivial advantage of phylogenetic methods is their ability to inform other levels of relationships (e.g., genus and family) and are not restricted to delimiting species. Multiple genome-based phylogenies have been constructed for taxonomic purposes (Garrity 2016; Hugenholtz et al. 2016; Yoon et al. 2017; Parks et al. 2018) and offer a more accurate resolution than 16S rRNA phylogenies (Brochier et al. 2005; Ciccarelli et al. 2006; Thiergart et al. 2014). Akin to sequence thresholds, phylogenetic approaches frequently rely on a single threshold (e.g., a phylogenetic distance) to define species, but recently, a new approach has been developed to reclassify all prokaryotic organisms, while correcting for the uneven evolutionary rates across the tree (Parks et al. 2018). Such approaches offer a universal framework to classify species—and other taxonomic ranks—across the Tree of Life, while correcting for uneven rates of evolution (i.e., defining species with lineage-specific thresholds). The application of these approaches is much more cumbersome than 16S and ANI thresholds, but online tools and resources to place newly sequenced genomes in a reference phylogenetic tree are now available (Parks et al. 2018). The development of such tools and the maintenance of online resources offer the possibility to classify all prokaryotic genomes with ease into a single phylogenetic framework. Although phylogenic methods offer many advantages over sequence threshold methods, they also require comprehensive taxon sampling and can be affected by the underlying phylogenetic model used to reconstruct the tree. Finally, a phylogenetic species concept is still based on ad hoc criteria and does not ambition to identify species based on an explicit speciation model.
The Stable Ecotype Model
The stable ecotype model (SEM) is a theoretical framework of bacterial evolution, upon which a microbial species concept can be founded (Cohan 2001; Wiedenbeck and Cohan 2011). In a world without sex, new beneficial alleles can only reach fixation through genome sweep (i.e., fixation of the entire genotype). Therefore, the competition of different bacterial strains for the same resources (the same niche) would lead periodically to the fixation of a single genotype. This model of periodic selection implies that most of the diversity of a species is periodically erased, thereby maintaining genetically cohesive entities, i.e., species. Thus, the SEM has the capacity to explain why bacteria form clusters of genomically similar entities. Under this framework, speciation is expected to occur when one strain gains the ability to colonize a different niche (Wiedenbeck and Cohan 2011). By colonizing a different niche, this new population would stop competing against the original population and would not be lost by the periodic selection of a successful genotype of the original population. Note that from the bacterial point of view, a new niche could be as simple as the presence of a new type of carbohydrate and multiple niches are expected to overlap in nature.
A theoretical difficulty of the SEM became apparent when comparing the gene content of bacteria. It became clear that the gene content of a single strain typically represents a very small fraction of the total gene repertoire of the species (i.e., the pangenome) (Tettelin et al. 2005; Medini et al. 2005; Vernikos et al. 2015). This implies that the genetic cohesion of microbial species is only true for a restricted fraction of their genes: their core-genome (Lapierre and Gogarten 2009). The scattered distribution of various accessory genes across strains sharing a highly conserved core-genome cannot be easily reconciled with the SEM. Although a substantial fraction of the pangenome corresponds to mobile elements (Bobay et al. 2013), accessory genes often contribute to the colonization of different niches (Ochman et al. 2000), which implies that the gain and losses of these genes can provide the capacity of a strain to colonize a new niche. This would lead to the disturbing conclusion that a given strain could frequently change species membership by gaining or losing specific sets of accessory genes. Because each genotype virtually contains its own set of accessory genes, each strain could be ascribed to a different ecotype and could be viewed as its own species (Doolittle and Zhaxybayeva 2009; Wiedenbeck and Cohan 2011). This extreme scenario, however, would fail to explain why many bacterial strains present a nearly identical core-genome.
Although the SEM does not easily accommodate the large diversity of accessory genes observed in related bacteria, it has been argued that the definition of an ecotype could be more flexible by encompassing multiple sub-niches (the “nano niche” model) (Wiedenbeck and Cohan 2011). Some strains of a community can acquire alleles or accessory genes specialized in a sub-niche, while remaining part of a broader ecologically-cohesive entity. These specialized strains within an ecotype can be perceived as new species in the making. Nascent speciation might be constantly occurring but need not lead to full speciation (Shapiro and Polz 2014) and this could potentially explain the vast pangenome diversity in bacterial species. Alternative mechanisms have been hypothesized to explain the extensive gene diversity within ecotypes such as a high turnover of accessory genes (Doolittle and Papke 2006) or ecological processes maintaining bacterial diversity such as phage predation (“kill the winner” hypothesis) (Rodriguez-Valera et al. 2009; Thingstad and Lignell 1997) or negative frequency-dependent selection (Cordero and Polz 2014).
While the SEM and related models could provide a coherent explanation of the observation of genomic clusters in the bacterial world—or at least their core-genomes—few results have reported genome sweeps as predicted by the periodic selection expected under the SEM. Multiple studies have overwhelmingly observed that gene sweeps rather than genome sweeps tend to occur under natural conditions (Simmons et al. 2008; Shapiro et al. 2012; Cadillot-Quiroz et al. 2012; Bendall et al. 2016). These results contradict one assumption made by the ecotype model: recombination is negligible relative to selection. Evidence of homologous recombination has been reported for the vast majority of analyzed prokaryotic species (Vos and Didelot 2009; Bobay and Ochman 2017a). That some evidence of homologous recombination exists for most species does not necessarily imply that the rates of homologous recombination are high enough to counteract genome sweeps. A more pertinent metric consists of comparing recombination rate relative to selection: the ratio r/s (Shapiro and Polz 2014). If selection is overwhelmingly strong relative to recombination, the selected genome is expected to reach fixation before the advantageous alleles are transferred to other genotypes. Because gene sweeps have been more frequently observed than genome sweeps in bacterial species, it seems that the relatively modest levels of homologous recombination in bacteria—in comparison to truly sexual organisms—would suffice to prevent genome sweeps unless extremely beneficial alleles are introduced.
Overall, the accumulation of empirical observations of gene sweeps in natural populations suggest that periodic selection might play a limited role in maintaining genomic cohesion in bacteria. Nevertheless, the SEM remains relevant for effectively clonal species (species with negligible rates of recombination), although the previously cited studies suggest that relatively few species might be effectively clonal (Vos and Didelot 2009; Bendall et al. 2016; Bobay and Ochman 2017a). An inherent difficulty of the SEM and other ecology-based definitions, in general, is the difficulty to gain accurate knowledge on microbial ecology and to identify what objective criteria can be used to define distinct niches. This lack of ecological data appears even more dramatic when compared to the colossal accumulation of genomic data. In the (meta-)genomic era, alternative approaches are needed. Starting from this observation, several authors have suggested the use of a reverse ecology approach, where, instead of searching for the genetic variants responsible for ecological segregation, it is more relevant to search for the ecological factors associated with allelic or accessory gene segregation (Shapiro and Polz 2014). The development of a reverse ecology framework potentially offers a powerful tool to extend our comprehension of the ecological factors driving the evolutionary dynamics and the cohesion of bacterial species.
Biological Species Concept
Sexual organisms engage in meiotic recombination at each generation and this maintains the genetic cohesion of species (Mayr 1942). The mechanisms leading to speciation in sexual organisms are diverse, can be either pre- or post-zygotic in nature, and are often conceptualized in the context of spatial arrangement of populations (sympatric or allopatric) (Coyne and Orr 2004; De Queiroz 2007). Most models assume that prolonged interruption of gene flow (e.g., zero or few migrants per generation) between two separated populations can lead to the independent accumulation of new alleles and new traits in each population through drift or local adaptation, leading to build up of reproductive incompatibilities and potentially triggering reinforcement, if the two populations are reunited. Other mechanisms, such as the appearance of incompatible alleles or alleles resulting in mating preferences, or even genomic duplications or rearrangements, can also lead to sexual barriers and, therefore, to the interruption of gene flow between populations. While evolution of reproductive barriers is often associated with speciation, it is important to realize that the interruption of gene flow can be either the cause or the consequence of speciation. In all scenarios, however, the interruption of significant gene flow remains associated with speciation, even if the barriers of gene flow can remain somewhat permissive after speciation (Mallet et al. 2007, 2016).
Although bacteria do not engage in true sexual reproduction, it has long been known that they are capable of exchanging DNA (Smith et al. 1993). Because gene flow is a common phenomenon across plants and animals as well as bacteria, this opens the possibility to define bacterial species with the same standards of the biological species concept (BSC) (Dykhuizen and Green 1991; Fraser et al. 2009; Bobay and Ochman 2017a). The fact that bacteria have the capacity to exchange DNA does not necessarily imply that they form biological species; instead, the real challenge is to determine whether the strength of gene flow is sufficient to shape cohesive bacterial units in bacteria, and thus whether common speciation models based on gene flow are applicable to bacteria as well. The question is then: how much and how frequently do they recombine? Can we detect these patterns of gene flow in bacteria as we do for sexual organisms? By “gene flow”, I exclusively refer to the replacement of DNA sequences by homologous recombination (also referred to as gene conversion). Homologous recombination consists of the exchange between two sequences of DNA that typically display a high identity in nucleotide composition (Vulic et al. 1997). In contrast to gene flow, horizontal gene transfer (HGT) refers to the gain of new genetic material without the replacement of a homologous sequence. This semantic differentiation allows for the distinction of gene segments of homologous genes that are exchanged (gene flow) versus new genes that are gained (HGT). Note that this distinction permits the differentiation of the outcome of the DNA transfer—homologous replacement or gain of DNA—but it does not necessarily involve different molecular mechanisms since HGT can involve homologous recombination between regions flanking the exchanged sequence (Mell et al. 2011; Croucher et al. 2012; Cordero et al. 2012; Everitt et al. 2014).
Two independent studies have scrutinized a relatively large range of prokaryotic species and came to the conclusion that a small proportion (<15%) of analyzed species do not show substantial signs of gene flow (Vos and Didelot 2009; Bobay and Ochman 2017a). In fact, similar numbers were estimated for viruses and there is growing evidence that the vast majority of cellular and acellular organisms engage in gene flow (Bobay and Ochman 2018a). In addition, many studies have reported that individual loci—rather than entire genotypes—sweep through natural populations (Simmons et al. 2008; Croucher et al. 2011; Shapiro et al. 2012; Cadillot-Quiroz et al. 2012; Bendall et al. 2016; Bao et al. 2016; Porter et al. 2017). These observations imply that gene flow is substantial enough to spread alleles—and even beneficial ones—to the entire population, suggesting the cohesive role of gene flow in bacterial genome dynamics. Importantly, the levels of gene flow across most bacterial species—and their variations—are often substantial enough to be detected using genomic datasets (Bobay and Ochman 2017a). Thanks to the vast accumulation of genomic data, it is possible to identify strains that do not engage in gene flow with the rest of the species (i.e., sexual isolation) by conducting large-scale resampling analyses. This allows to classify sexual eukaryotes, bacteria, archaea, and even viruses under a unique BSC-based species definition.
The delimitation of species based on gene flow is more cumbersome than ANI sequence thresholds, since it requires identification of the core-genome (or a portion thereof) for the tested genome sample and estimation of distances or tree topologies and potentially conducting resampling analyses (Bobay and Ochman 2017b). Similar to phylogenetic methods, it is also possible to compare individual genomes to a database of preprocessed species available online (i.e., ConSpeciFix) (Bobay et al. 2018), which facilitates the classification of newly sequenced data. Detecting and quantifying gene flow remains a delicate endeavor as evidenced by the lack of a consensual methodology to infer homologous recombination. Various methods to estimate recombination rates exist, but they often rely on different models and assumptions regarding the recombination process (Didelot and Falush 2007; Marttinen et al. 2012; Yahara et al. 2014, 2015; Didelot and Wilson 2015; Mostowy et al. 2017), and this contributes to the inference of inconsistent estimates of recombination rates across studies (Bobay et al. 2015). Recently, we introduced a methodology based on the quantification of homoplasies to detect gene flow across large genomic datasets (Bobay and Ochman 2017a; Bobay et al. 2018). Homoplasies are polymorphisms incompatible with vertical inheritance from a shared ancestor and are mostly introduced by gene flow (Bobay and Ochman 2017a). Although the ratio between homoplasic and non-homoplasic polymorphisms does not provide an accurate metric to quantify recombination rates, the detection of homoplasies is rather straightforward and does not rely on complex model assumptions and over parametrization. Interestingly, this homoplasy-based approach appears more robust to genome resampling and gene bootstrapping when compared to ClonalFrameML (Bobay and Ochman 2018b). Inferring gene flow based on homoplasies is limited to the detection of recombination events internal to the dataset and the method does not aim to model imports from external sources. Recombining species can sometimes be misclassified as clonal when multiple sexually isolated genomes are included in the analysis and the sample size is too small to resample and test subpopulations for gene flow; thus, the method is most efficient when large datasets are available and when genetic diversity is high. This limitation will be resolved as more genomes will be sequenced, but, to this date, the analysis of several species can remain inconclusive due to ambiguous signals (Bobay and Ochman 2017a). In addition, the recent accumulation of metagenomic data combined with the development of bioinformatics tools that resolve strain genotypes within metagenomic samples (Nayfach et al. 2016; Pasolli et al. 2017; Truong et al. 2017) constitutes a new source of data readily exploitable to define species based on gene flow.
Because bacteria can sometimes gain genes from other species through HGT, it has been argued that bacteria might not fit a BSC definition in comparison to truly sexual organisms. Species borders are somewhat “fuzzy” for bacteria (Hanage et al. 2005; Hanage 2013) and many studies have detected HGT events in prokaryotes, leading to the conclusion that they might be genomically promiscuous (Popa and Dagan 2011). It should be emphasized, however, that gene flow between species remains very rare when considering the overall time scale of prokaryote evolution, and HGT events occur primarily between related bacteria (Popa et al. 2011). In contrast, gene flow within species is expected to occur at much higher frequencies relative to the acquisition of new genes from external species by HGT (Caro-Quintero et al. 2009; Cadillot-Quiroz et al. 2012; Shapiro et al. 2012; Krause and Whitaker 2015; David et al. 2017). Comparison of ~100 species indicates that most bacteria show clear signs of gene flow and the same method can also retrieve species borders in well classified animals such as humans and Drosophila (Bobay and Ochman 2017a). It is well established that sexual eukaryotes are not as well isolated as previously thought (Danchin and Rosso 2012; Syvanen 2012), but introgression and incomplete lineage sorting do not typically prevent defining species borders in truly sexual organisms (Mallet et al. 2016). Although eukaryotic and prokaryotic species borders can be “leaky” and occasionally allow gene flow from external sources, this process need not be prevalent enough to blur species borders (Mallet 2008).
Given the commonality of genomic exchange across diverse types of organisms, a BSC-based definition allows the use of a universal species concept to classify all lifeforms under a biologically relevant definition. What are the implications of applying such a species concept to microbes? Most BSC-species (i.e., bacterial species classified based on the BSC) correspond to closely related genomes that typically present ≥95% ANI (Bobay and Ochman 2017a). However, this is not always true since several BSC-species contain genomes that would not be classified as members of the same species based on ANI thresholds and, conversely, other BSC-species were found to exclude members that would be part of the same species according to ANI thresholds (≥95% ANI) (Bobay and Ochman 2017a). These results are in agreement with analyses showing that a single ANI or phylogenetic threshold fails to define consistent species across prokaryotes (Parks et al. 2018; Wright and Baum 2018). These differences can be putatively ascribed to the use of more-or-less permissive recombination mechanisms across species. Experimental data have suggested that the frequency of homologous recombination decreases exponentially with sequence divergence (Roberts and Cohan 1993; Zawadzki et al. 1995; Vulic et al. 1997; Majewski and Cohan 1998; Majewski et al. 2000) due to the action of the mismatch repair system (Matic et al. 2000). These observations suggest a simple model of sexual isolation in bacteria. The action of the mismatch repair system seems highly variable across taxa (Majewski 2001), which suggests that barriers of gene flow driven by sequence divergence would also be variable across species. In contrast to these observations, there is no systematic negative correlation between recombination and sequence divergence (Bobay and Ochman 2017a) and gene flow has been reported between bacteria presenting relatively divergent genomes (Sheppard et al. 2008; Mell et al. 2011; Cordero et al. 2012), suggesting that sequence divergence plays a limited role in establishing barriers of gene flow. These discrepancies between experimental data and genome analyses can be explained by multiple factors. Firstly, gene flow is detected by the exchange of polymorphisms, and recombination events that do not result in any exchange of polymorphisms can remain invisible to some approaches. This implies that the rates of recombination between highly similar genomes are frequently underestimated. Secondly, selection can potentially have a strong impact in selecting—positively or negatively—alleles exchanged by gene flow, mirroring adaptive introgression or Dobzhansky–Muller incompatibilities in sexual organisms (Mallet et al. 2016). Finally, a simpler explanation might account for these discrepancies. The exponential relationship between sequence identity and recombination rate is based on the observation that nearly identical regions flanking the recombination tract—the minimum efficiently processed segments (MEPS)—are needed to initiate recombination (Shen and Huang 1986; Wiedenbeck and Cohan 2011; Hanage 2016). However, sequence identity need not be high along the entire segment of recombined DNA because recombination requires high sequence identity only along the MEPS, which are only ~26 nt long (Shen and Huang 1986; Wiedenbeck and Cohan 2011; Hanage 2016). This suggests that more variable sequences of DNA might be exchanged as long as a few clusters of nearly identical nucleotides remain available to initiate homologous recombination.
Mixed Model
The SEM and a BSC-like model of bacterial evolution need not be fundamentally opposed. A BSC-like model is, by definition, unable to define species borders for clonal species. It is also likely that species with low rates of recombination would appear effectively clonal when analyzing genomic data, meaning that the BSC will fail to accurately delimit species in some bacterial groups. For these clades, the SEM appears the most pertinent force maintaining genetic cohesion and therefore is most appropriate to define the borders of these species. The fact that very few studies have reported genome sweeps relative to gene sweeps suggests the prevalence and significance of recombination in bacteria and implies that the vast majority of bacterial species can be defined based on the BSC. Both models could, therefore, be integrated to define species; the SEM for lineages that are effectively clonal and a BSC-like model for species that appear effectively sexual. A key distinction between both models is that the SEM is inherently ecologically centered, whereas a BSC-based model of bacterial evolution does not necessarily involve ecological mechanisms. However, the speciation processes through new niche colonization assumed under the SEM can also lead to speciation under the BSC.
3 Speciation: From Maintenance to Disruption of Genomic Cohesion
Neutral Processes
Simulations have provided insightful answers regarding the impact of neutral evolution on the formation of new species. In the absence of recombination, it is expected that some distinct genome clusters would emerge in sympatry (Fraser et al. 2007). However, most of these newly emerged clusters are expected to go extinct through drift. On the other hand, gene flow allows populations to maintain cohesive genomes (Fraser et al. 2007; Friedman et al. 2013). These results suggest that neutral evolution is unlikely to promote the emergence of new species in bacteria, especially in the case of recombining populations. It has been noted that this neutral model of speciation does not consider the potential barrier of gene flow imposed by sequence divergence (Fraser et al. 2007), in which case, it may be possible that divergent genome clusters become more and more sexually isolated. It should be underlined, however, that neutral evolution is expected to drive divergence very slowly, and due to the frequent loss of newly emerged clusters by drift, it is unlikely that population clusters would accumulate enough mutations to impose a substantial barrier of gene flow.
Geography
The previous model of neutral speciation has been developed for sympatric populations (i.e., geographically overlapping populations), which is thought to be the preponderant situation in bacteria (Vos 2011; Shapiro and Polz 2015). However, geographic differentiation suggests that allopatric speciation could occur in bacteria (Simmons et al. 2008; Denef et al. 2010; Whitaker et al. 2003; Reno et al. 2009; Krause and Whitaker 2015). Processes resembling allopatric speciation with the interruption of gene flow in bacteriophages targeting different receptors have even been observed in an experimental evolution setting (Meyer et al. 2016). The impact of geography remains elusive since species spanning large continental and oceanic distributions can remain genetically cohesive (Papke et al. 2007; Coleman and Chisholm 2010; Boucher et al. 2011). Recent modeling work has emphasized the impact of niche overlap in bacterial speciation, further revealing the importance of habitat structure in promoting genomic isolation, especially for recombining bacteria (Marttinen and Hanage 2017). The spatial dynamics of microbial distributions remains difficult to characterize and seemingly overlapping populations might not necessarily encounter each other due to fine-scale habitat structure (i.e., mosaic sympatry) (Mallet 2008; Shapiro and Polz 2014).
Recombination Barriers
As mentioned above, the initiation of homologous recombination requires the presence of nearly identical short sequences (i.e., MEPS) (Vulic et al. 1997; Majewski and Cohan 1999) and, although relatively divergent sequences can engage in gene flow, sequence divergence can affect recombination rates due to the frequency of available MEPS to initiate recombination. Interestingly, the sequence (MEPS) conservation required to initiate recombination seems to be dependent on the mismatch repair (MMR) system (Matic et al. 2000), which can be more or less permissive across species and strains. The evolution—and sometimes the complete loss—of the MMR system is therefore expected to have a strong impact on sexual isolation in prokaryotes.
Restriction–Modification (RM) systems are frequently used by bacteria to protect themselves against mobile elements and, in particular, bacteriophages (Thomas and Nielsen 2005; Labrie et al. 2010). The presence of different RM systems across strains and species can lead to incompatibilities of gene flow and this has been found to regulate and structure gene flow (Oliveira et al. 2014, 2016). Consequently, the gain or loss of RM systems can have direct consequences on the interruption of gene flow and can potentially lead to speciation. In theory, CRISPR–Cas systems might exhibit similar properties, but since they specifically target a limited number of sequences, they are unlikely to introduce genome-wide incompatibilities. Because of these properties, RM systems can shape the networks of gene flow and the population structure of bacterial species. These systems might drive the establishment of durable barriers of gene flow, potentially leading to speciation.
Gene flow relies on the presence of different vectors and mechanisms capable of disseminating and capturing DNA. The three main mechanisms of DNA transfer, namely transformation, conjugation, and transduction, present diverse degrees of specificity. (i) Transformation does not require cell–to–cell interactions, since environmental DNA is directly taken up by the cell; but recipient cells need to be competent, and relatively few bacteria are known to naturally engage in this process (Johnston et al. 2014). Some bacteria engaging in transformation such as Neisseria and Pasteurellaceae require the presence of specific DNA uptake sequences or uptake signal sequences (Goodman and Scocca 1988; Scocca et al. 1974; Danner et al. 1982), thereby restricting the range of potential DNA donors to related lineages. Moreover, due to the rapid degradation of DNA when released in the environment this mechanism likely requires close proximity between cells, suggesting that transformation might only mediate gene flow between sympatric populations. (ii) Conjugation involves more constrained transfers of DNA through cell–to–cell contacts, which is mediated by specific pilus interactions and type IV secretion systems (Guglielmini et al. 2013). These conjugative transfers occur primarily between conspecifics, although plasmids have been shown to be occasionally exchanged across much more divergent lineages (Smillie et al. 2010). Because this process requires the direct contact of cells, gene flow mediated by the conjugative apparatus must also occur in sympatry. (iii) Transduction is another route for gene flow where bacterial DNA is packaged within phage particles or gene transfer agents (GTAs) (Lang and Beatty 2007; Popa and Dagan 2011). Phage particles are rarely able to infect multiple species and are often restricted to a subset of strains (Popa et al. 2017). As opposed to transformation and conjugation, phage particles can potentially transport DNA over longer distances (and potentially for long periods of time), suggesting that allopatric—and perhaps anachronistic—populations are able to engage in some levels of gene flow without requiring migration. These three mechanisms, and especially conjugation and transduction, rely on specific molecular signals and are typically restricted to conspecific cells. The overall specificity of these mechanisms is expected to favor gene flow within species rather than between species. Conjugation and transduction also potentially have important consequences for bacterial speciation, since the loss of cell-vector specificity can lead to the partial or complete interruption of gene flow.
Selection
As mentioned above, neutral processes are unlikely to lead to bacterial speciation, especially in the case of sympatric recombining populations that co-occur at fine spatial scales (Fraser et al. 2007). This suggests that selection must initiate the formation of distinct genomic clusters, which might eventually lead to selection against genetic intermediates and the cessation of gene flow (Shapiro 2014). Ecological specialization is thought to be a strong force leading to speciation, since the nascent species will present differentially selected EcoSNPs or specialized accessory genes, i.e., alleles or genes specialized in one niche (Shapiro et al. 2012). Simulations have shown that sympatric speciation is more likely when fewer loci are required for speciation and when recombination is reduced (Friedman et al. 2013). As two populations become more and more differentiated, the accumulation of substitutions is expected to reduce gene flow due to epistatic interference (Jain et al. 1999), similarly to Dobzhansky–Muller incompatibilities. Indeed, many loci of the genome coevolve together, and, for instance, central protein complexes such as translation, transcription, and replication complexes require interaction between many central proteins that coevolved together, which could explain why these genes are rarely exchanged by HGT across species, i.e., the “complexity hypothesis” (Jain et al. 1999). Such incompatibilities are expected to be most relevant when populations have significantly diverged and most likely form barriers of gene flow when DNA originates from distant species. However, it is possible that those negatively selected epistatic interactions also contribute to the isolation of more recently diverged populations.
Several studies have demonstrated that the impact of selection on bacterial genome evolution depends on the relative prevalence of selection (s) and recombination rate (r) in sympatric evolution (Shapiro et al. 2009; Friedman et al. 2013; Polz et al. 2013). When selection is much stronger than recombination (r/s << 1), the selected allele will lead to the fixation of the entire genotype through genome sweep. The resulting process will be similar to the periodic selection predicted by the SEM. On the other hand, alleles with lower selective coefficients relative to recombination (r/s >> 1) are expected to evolve by gene/allele sweep. In this case, selection will be unable to lead to speciation as the selected allele will be exchanged between the population’s genotypes by gene sweep. Several studies have attempted to determine whether prokaryotic populations evolve primarily through gene or genome sweeps and, so far, evidence overwhelmingly suggests that gene sweeps are more frequent than genome sweeps (a single case of genome sweep against ~35 cases of gene sweeps (Simmons et al. 2008; Croucher et al. 2011; Shapiro et al. 2012; Cadillot-Quiroz et al. 2012; Bendall et al. 2016; Bao et al. 2016; Porter et al. 2017)). The large prevalence of gene sweeps over genome sweeps is somewhat surprising considering that prokaryotes, as asexual organisms, are thought to display modest rates of gene flow (Wiedenbeck and Cohan 2011). It is, however, difficult to clearly quantify the impact of gene flow on genome evolution (Bobay et al. 2015) and a recent experimental evolution study has shown that gene flow can even lead to the extinction of beneficial alleles (Maddamsetti and Lenski 2018). It is possible that additional factors counteract genome sweeps, such as clonal interference (Lieberman et al. 2014; Maddamsetti et al. 2015) and negative frequency-dependent selection (Cordero and Polz 2014; Takeuchi et al. 2015).
Introgression and HGT from External Species
In comparison to the processes acting in sexual organisms, occasional gene flow from external bacteria could be seen as a form of introgression. It has been noted that introgression can sometimes present a source of adaptive alleles in sexual organisms and those transfers can even lead to hybrid speciation (Mallet 2007; Rieseberg 1997; Seehausen 2004; Keller et al. 2013). The importance of these processes remains to be explored in prokaryotes. A study comparing the evolution of two Campylobacter species—C. jejuni and C. coli—can be viewed as evidence of bacterial introgression (Sheppard et al. 2008, 2013). Although these results might lead to the complete “despeciation” of the two lineages, it should be noted that the transfer of DNA is asymmetric where one clade of C. coli has likely gained alleles from C. jejuni but other clades of C. coli did not. Interestingly, this case of bacterial introgression appears ecologically-driven based on recent niche overlap (Sheppard et al. 2008). It is, therefore, possible that introgression can result in the same outcomes in prokaryotes, such as hybrid speciation (Shapiro et al. 2016).
Similar to introgression, the gain of new genes from distinct species by HGT offers another means to colonize new niches through ecologically-driven adaptation. The acquisition of antibiotic-resistant genes constitutes a well-documented case, but many other examples have been reported (Ochman et al. 2000; Popa and Dagan 2011). It has been shown that HGT—rather than duplication—plays a predominant role in introducing new paralogs in the pangenome of prokaryotic species (Treangen and Rocha 2011), although these genes frequently come from related species due to genetic incompatibilities (i.e., gene promoters/regulators and codon usage bias) (Sorek et al. 2007; Popa et al. 2017). These acquired genes can mediate the colonization of new niches and can potentially lead to ecology-driven speciation. However, as noted above, accessory genes are not stably associated with a given genotype and tend to be frequently exchanged across strains of a given species (Schubert et al. 2009), indicating that they do not necessarily drive the formation of distinct ecologically specialized entities (Shapiro and Polz 2015).
Summary
Across the many forces that can affect speciation, it should be noted that neutral processes such as population dynamics and sequence divergence are unlikely to lead to speciation in bacteria, and that selection seems to be a necessary force by initiating and maintaining speciation. Selection in bacteria can act through two predominant avenues: (i) by driving ecological adaptation to different niches following, for instance, the gain of new genetic material and (ii) by preventing gene flow between populations due to the presence of genetic incompatibilities, such as different RM systems, vector specificity, or negative epistasis. Other factors such as population dynamics and geographic range have been found to have an impact on speciation, although their relative contribution remains to be precisely deciphered. Overall, a BSC-based speciation model in prokaryotes would also rely on ecological processes and selection, as hypothesized by the SEM. However, one major difference with the SEM is that a BSC-based model of prokaryotic speciation predicts that speciation events can be driven by genetic incompatibilities and need not be systematically adaptive and ecologically-driven.
4 Species Borders and Pangenome Borders
Pangenome and Species Definitions
The definition of species has direct consequences regarding the definition of pangenomes. If bacterial species are defined based on inconsistent criteria, it is not possible to compare the size of the pangenome across species and lineages. The case of Prochlorococcus illustrates this issue particularly well. Prochlorococcus is often studied as a single entity since it constitutes a single species based on 16S rRNA thresholds but multiple species based on ANI thresholds. The pangenome of Prochlorococcus has been estimated to reach the impressive amount of ~75,000 genes (Kashtan et al. 2014), although this would include strains that present less than 70% ANI, and this entity would actually correspond to multiple species and even genera. This issue likely affects many pangenome analyses considering that public databases frequently contain misclassified species and species classified based on inconsistent methods (Martiny et al. 2006; Comas et al. 2009; Trost et al. 2010). Studies focusing on the evolution of bacterial pangenomes should be based on rigorous species delimitation, since the misclassification of a single genome can lead to dramatic overestimates or underestimates of the size of a species’ pangenome.
Species delimitation is not the only concern when analyzing pangenomes. The number of genomes sampled for each species obviously impacts pangenome estimates, since pangenomes necessarily increase in size as more genomes are included. It is possible to test if pangenome size reaches a plateau by performing resampling analyses, which would indicate that a sufficient number of genomes have been sampled to estimate the true pangenome size of the analyzed species (Tettelin et al. 2005; Lapierre and Gogarten 2009). Alternatively, it is possible to apply resampling analyses or to correct these metrics to account for uneven sampling biases across species (Bobay and Ochman 2018b). Biases in species sampling are a common issue for many genomic analyses and several methods have been developed as an attempt to address this shortcoming (Lapierre et al. 2016). However, the most efficient solution remains to increase sample sizes, and, more importantly, to limit biases when collecting samples, but this last consideration is often in conflict with study designs focusing on medically- or environmentally-relevant strains.
Cohesion of Core- and Pangenomes
The goal of a species definition is to identify cohesive ensembles of evolutionary lineages. The ideal species definition would succeed in identifying genetically and ecologically cohesive units. Although genetic cohesion is easier to assess than ecological cohesion for bacteria, the genetic homogeneity of a group of organisms can be evaluated through different lenses. Firstly, because the core-genome constitutes the backbone of genes shared by all members of the species, these genes are more readily used to infer evolutionary relatedness and other metrics. Moreover, despite gene flow, core-genomes have conserved the phylogenetic signal of the vertical inheritance of bacterial taxa (Touchon et al. 2009; Abby et al. 2012). Nearly all genome-based species definitions—i.e., ANI, phylogenetic methods, and BSC-like—rely exclusively on the cohesion of the core-genome. The pangenome potentially offers an alternative measure of the genetic cohesion of species, since conspecific strains are expected to share more similar gene repertoires than strains belonging to distinct species. It is currently difficult to assess the pangenome cohesion of a species considering that accessory genes tend to be found at low frequency within species and this would require deep genome sampling, although more and more bacterial species have now hundreds or thousands of sequenced genomes. More analyses need to be performed to understand the specificity of pangenomes, especially in relation to closely related lineages and ecologically or geographically overlapping species.
Gene flow can define biological species based on DNA exchange along the core-genome but, so far, this method has been ignoring the patterns of HGT of the pangenome. The core- and pangenomes are two complementary metrics that can be used to infer the cohesion of species and some recent results obtained in two bacterial phyla suggest that core- and pangenomes present the same phylogenetic signal, implying that both can be reliable for inferring species borders (Wright and Baum 2018). In fact, a recent method has proposed a first attempt to delimitate species based on pangenome cohesion (Moldovan and Gelfand 2018), which opens promising possibilities to include pangenome cohesion into species delimitation. More work needs to be done in order to finely understand the evolutionary dynamics of the pangenome itself. For instance, the dynamics of the pangenome is likely affected by the ability of a given species to engage in gene flow, as suggested by a study showing that clonal species are unlikely to present a large pangenome, since their pangenome primarily evolves through gene loss (Bolotin and Hershberg 2015). Bacterial species can also gain new genes from external lineages and the extent of segregation of the pangenome remains poorly understood. The accumulation of genomic data should soon allow more accurate analysis of the dynamics of the pangenome and this will open new avenues for evaluating the genetic cohesion of prokaryotic species.
5 Drift-Barrier Model for Pangenome Evolution
A BSC-based species definition is particularly relevant for studying population genetics in prokaryotic organisms. Several parameters such as recombination rate, effective population size (Ne), or pangenome size are metrics that are typically inferred at the species level. In particular, Ne has strong implications regarding the relative impact of selection and drift acting on a given species. High Ne populations are less sensitive to drift and can efficiently purge deleterious sequences, whereas low Ne populations, on the other hand, will not be as effective at purging deleterious mutations. A trait conferred by a given variant would primarily evolve through drift (i.e., neutrally) when |2.Ne.s| << 1, while selection will be effective when |2.Ne.s| >> 1, where s represents the selection coefficient of a given sequence or variant (Kimura 1968). For these reasons, it is believed that more complex organisms such as mammals, which have low Ne, present larger genomes due to the accumulation of “junk DNA” through drift (i.e., the Mutational Hazard Hypothesis) (Lynch and Conery 2003; Lynch et al. 2011). Because these organisms display small population sizes, selection is not as efficient at purging slightly deleterious sequences, such as noncoding DNA, introns, and mobile elements.
In contrast to many eukaryotes, bacterial genomes are small and compact and because microbes present much larger population sizes, this seems in perfect agreement with the expectation of the Mutational Hazard hypothesis. The genomic compactness of bacteria has been ascribed to a strong bias toward deletion in these organisms (Mira et al. 2001; Andersson and Andersson 2001). However, several studies have observed that, across bacteria, genome size appears positively correlated with Ne (Daubin and Moran 2004; Kuo et al. 2009; Novichkov et al. 2009). Free-living bacteria frequently possess relatively large genomes (typically >3 Mb), while obligate endosymbionts—with low Ne—have smaller genomes (frequently <1 Mb) (Moran and Plague 2004). Yet, some marine bacteria, which are thought to reach gigantic population sizes, also present streamlined genomes (Giovannoni et al. 2005, 2014). In particular, Prochlorococcus and Pelagibacter ubique have small genomes (~1 Mb), although they might be among the most abundant cellular organisms on earth (Batut et al. 2014). Therefore, the relationship between Ne and genome size appears to be more complex in bacteria.
One key difference between bacteria and higher eukaryotes is the very low amount of noncoding DNA, introns and mobile elements found in most bacterial genomes. In prokaryotes, variations in genome size are primarily driven by the presence of different amounts of accessory genes. Accessory genes are assumed to be functional and beneficial to the cell and recent modelling work suggests that virtually all genes in prokaryotic genomes are expected to be beneficial (Sela et al. 2016). Because the diversity of accessory genes is a direct function of pangenome size, this opens the possibility that Ne may drive the evolution of pangenome size rather than average genome size in prokaryotes. In support to this hypothesis, clear correlations between Ne and pangenome size have been observed across a dataset of 153 species, whose borders have been defined based on the BSC under a unified framework (Bobay and Ochman 2018b). Other recent studies have also reported similar trends (Mcinerney et al. 2017; Andreani et al. 2017).
Based on these observations, we have recently proposed that bacterial pangenomes could be driven by Drift-Barrier evolution (Bobay and Ochman 2018b). The Drift-Barrier model has originally been developed to account for the variation in mutation rates across organisms (Sung et al. 2012; Lynch et al. 2016). Under a Drift-Barrier model, pangenome size is expected to be a function of Ne because only the most beneficial accessory genes would be conserved by selection in small Ne species, while species with large Ne would be able to conserve accessory genes with modest fitness contribution (Fig. 1). As supported by multiple studies, deleterious and neutral sequences are expected to be quickly purged from microbial genomes (Mira et al. 2001; Andersson and Andersson 2001). Our model assumes that virtually every gene of the pangenome is beneficial (positive selection coefficient: s > 0). Even if beneficial, an accessory gene is expected to be retained by selection only if it is perceived as effectively beneficial. In other words, an accessory gene will be conserved when 2.Ne.s >> 1, while genes that appear effectively neutral (2.Ne.s << 1) are expected to be lost by drift. This implies that high Ne species are expected to retain a larger pool of genes including many accessory genes with modest fitness contribution, whereas low Ne species can only conserve the most beneficial genes (high s), i.e., mostly essential and/or core genes. Although new genes can be introduced into a species’ pangenome by HGT, those accessory genes with low selective coefficient will be lost by drift.
Drift-Barrier model of pangenome evolution. Each large circle represents a pangenome and small circles represent individual genes. Color gradient reflects the selective coefficient of the genes. Species with large effective population size Ne are less subject to drift and can retain genes of small beneficial value (left). As Ne decreases, additional genes of small fitness benefit will be perceived as effectively neutral and will be lost by drift (center). Under strong levels of drift, as expected in small Ne species, only the most beneficial genes will be conserved by selection, and this will result in small pangenomes mostly composed of core/housekeeping genes (right)
6 Outlook
Many aspects of bacterial biology are now better understood but building a biologically-relevant microbial species concept remains challenging. Because prokaryotic organisms are microscopic, their population dynamics, ecological interactions, and speciation mechanisms are still difficult to decipher. Many aspects of the population processes driving microbial evolution have not been characterized. Habitat structure—and its temporal variations—of prokaryotic species is still for the large part mysterious. Similarly, microbial ecology and its impact on population dynamics remain tedious to describe in depth. Defining clear microbial niches is problematic practically and conceptually and little is known about microbial ecology compared to the vast collection of genomic data now available. The recent development of reverse ecology approaches opens a new route to gain knowledge about microbial ecology.
The accumulation of genomic data has profoundly impacted our vision of speciation in prokaryotic organisms. Several results suggest that prokaryotic species are definable and diagnosable as genetically cohesive as evidenced by the existence of a core-genome. However, the evolution of the core-genome remains to be fully understood. It is becoming possible to analyze the evolution of species- and genus-specific core-genomes over relatively short evolutionary time scales by comparing related species when sufficient genomic data is available (Touchon et al. 2014). On the other hand, the vast diversity of microbial pangenomes emphasizes the versatility of bacterial species. Much larger data sets are needed to accurately understand the dynamics of bacterial pangenomes, but several species now have thousands of sequenced genomes available. Deciphering the evolution of the pangenome will be highly insightful for our understanding of the dynamics and the genomic cohesion of microbial species.
From the original view of bacteria as purely clonal organisms, more and more evidence indicate that gene flow and HGT are key players in the evolution of most bacteria, and potentially act as major contributors to bacterial speciation. Computational approaches are needed to finely characterize gene flow in order to understand how networks of DNA routes can drive genomic cohesion and division in microbial species. Integrating these different aspects of bacterial biology will contribute to a more comprehensive prokaryotic species concept.
References
Abby SS, Tannier E, Gouy M, Daubin V (2012) Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci U S A 109:4962–4967
Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF (2004) Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186:2629–2635
Andersson JO, Andersson SG (2001) Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol Biol Evol 18:829–839
Andreani NA, Hesse E, Vos M (2017) Prokaryote genome fluidity is dependent on effective population size. ISME J 11(7):1719
Avery OT, MacLeod MC, McCarty M (1944) Studies on the chemical nature of the substance inducing transformation of pneumonococcal types. Induction of transformation by a deoxyribo-nucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–157
Bao YJ, Shapiro BJ, Lee SW, Ploplis VA, Castellino FJ (2016) Phenotypic differentiation of Streptococcus pyogenes populations is induced by recombination-driven gene-specific sweeps. Sci Rep 6:36644
Bapteste E, O’malley MA, Beiko RG, Ereshefsky M, Gogarten JP, Franklin-Hall L, Lapointe FJ, Dupre J, Dagan T, Boucher Y, Martin W (2009) Prokaryotic evolution and the tree of life are two different things. Biol Direct 4:34
Batut B, Knibbe C, Marais G, Daubin V (2014) Reductive genome evolution at both ends of the bacterial population size spectrum. Nat Rev Microbiol 12:841–850
Bendall ML, Stevens SL, Chan LK, Malfatti S, Schwientek P, Tremblay J, Schackwitz W, Martin J, Pati A, Bushnell B, Froula J, Kang D, Tringe SG, Bertilsson S, Moran MA, Shade A, Newton RJ, Mcmahon KD, Malmstrom RR (2016) Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J 10:1589–1601
Bobay LM, Ochman H (2017a) Biological species are universal across life’s domains. Genome Biol Evol 9:491–501
Bobay LM, Ochman H (2017b) Impact of recombination on the base composition of bacteria and archaea. Mol Biol Evol 34:2627–2636
Bobay LM, Ochman H (2018a) Biological species in the viral world. Proc Natl Acad Sci U S A 115:6040–6045
Bobay LM, Ochman H (2018b) Factors driving effective population size and pan-genome evolution in bacteria. BMC Evol Biol 18:153
Bobay LM, Rocha EP, Touchon M (2013) The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol 30:737–751
Bobay LM, Traverse CC, Ochman H (2015) Impermanence of bacterial clones. Proc Natl Acad Sci U S A 112:8893–8900
Bobay LM, Ellis BS, Ochman H (2018) ConSpeciFix: classifying prokaryotic species based on gene flow. Bioinformatics 21:3738–3740
Bolotin E, Hershberg R (2015) Gene loss dominates as a source of genetic variation within clonal pathogenic bacterial species. Genome Biol Evol 7:2173–2187
Boucher Y, Cordero OX, Takemura A, Hunt DE, Schliep K, Bapteste E, Lopez P, Tarr CL, Polz MF (2011) Local mobile gene pools rapidly cross species boundaries to create endemicity within global Vibrio cholerae populations. MBio 2:e00335
Brenner DJ, Staley J, Krieg N (2000) Classification of prokaryotic organisms and the concept of bacterial speciation. In: Boone DR, Castenholz RW, Garrity GM (eds) Bergey’s manual of systematic biology, 2nd edn. Springer, New York
Brochier C, Forterre P, Gribaldo S (2005) An emerging phylogenetic core of archaea: phylogenies of transcription and translation machineries converge following addition of new genome sequences. BMC Evol Biol 5:36
Buchrieser C, Glaser P, Rusniok C, Nedjari H, D’hauteville H, Kunst F, Sansonetti P, Parsot C (2000) The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 38:760–771
Cadillot-Quiroz H, Didelot X, Held N, Herrera A, Darling A, Reno M, Krause DJ, Whitaker RJ (2012) Patterns of gene flow define species of Thermophilic Archaea. PLoS Biol 10:e1001265
Caro-Quintero A, Konstantinidis KT (2012) Bacterial species may exist, metagenomics reveal. Environ Microbiol 14:347–355
Caro-Quintero A, Rodriguez-Castano GP, Konstantinidis KT (2009) Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species. J Bacteriol 191:5824–5831
Ciccarelli FD, Doerks T, Von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287
Cohan FM (2001) Bacterial species and speciation. Syst Biol 50:513–524
Coleman ML, Chisholm SW (2010) Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci U S A 107:18634–18639
Comas I, Homolka S, Niemann S, Gagneux S (2009) Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815
Cordero OX, Polz MF (2014) Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12:263–273
Cordero OX, Ventouras LA, Delong EF, Polz MF (2012) Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci U S A 109:20059–20064
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland, MA
Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, Van Der Linden M, Mcgee L, Von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434
Croucher NJ, Harris SR, Barquist L, Parkhill J, Bentley SD (2012) A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog 8:e1002745
Danchin EG, Rosso MN (2012) Lateral gene transfers have polished animal genomes: lessons from nematodes. Front Cell Infect Microbiol 2:27
Danner DB, Smith HO, Narang SA (1982) Construction of DNA recognition sites active in Haemophilus transformation. Proc Natl Acad Sci U S A 79:2393–2397
Daubin V, Moran NA (2004) Comment on “the origins of genome complexity”. Science 306:978; author reply 978
David S, Sanchez-Buso L, Harris SR, Marttinen P, Rusniok C, Buchrieser C, Harrison TG, Parkhill J (2017) Dynamics and impact of homologous recombination on the evolution of Legionella pneumophila. PLoS Genet 13:e1006855
De Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886
De Queiroz K, Gauthier J (1994) Toward a phylogenetic system of biological nomenclature. Trends Ecol Evol 9:27–31
Denef VJ, Mueller RS, Banfield JF (2010) AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J 4:599–610
Didelot X, Falush D (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266
Didelot X, Wilson DJ (2015) ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041
Dobzhansky T (1935) A critique of the species concept in biology. Philos Sci 2:344–355
Doolittle WF, Papke RT (2006) Genomics and the bacterial species problem. Genome Biol 7:116
Doolittle WF, Zhaxybayeva O (2009) On the origin of prokaryotic species. Genome Res 19:744–756
Dorer MS, Sessler TH, Salama NR (2011) Recombination and DNA repair in Helicobacter pylori. Annu Rev Microbiol 65:329–348
Dykhuizen DE, Green L (1991) Recombination in Escherichia coli and the definition of biological species. J Bacteriol 173:7257–7268
Escobar-Paramo P, Giudicelli C, Parsot C, Denamur E (2003) The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J Mol Evol 57:140–148
Everitt RG, Didelot X, Batty EM, Miller RR, Knox K, Young BC, Bowden R, Auton A, Votintseva A, Larner-Svensson H, Charlesworth J, Golubchik T, Ip CL, Godwin H, Fung R, Peto TE, Walker AS, Crook DW, Wilson DJ (2014) Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat Commun 5:3956
Fraser C, Hanage WP, Spratt BG (2007) Recombination and the nature of bacterial speciation. Science 315:476–480
Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP (2009) The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741–746
Friedman J, Alm EJ, Shapiro BJ (2013) Sympatric speciation: when is it possible in bacteria? PLoS One 8:e53539
Garrity GM (2016) A new genomics-driven taxonomy of bacteria and archaea: are we there yet? J Clin Microbiol 54:1956–1963
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ (2005) Genome streamlining in a cosmopolitan oceanic bacterium. Science 309(5738):1242–1245
Giovannoni SJ, Cameron Thrash J, Temperton B (2014) Implications of streamlining theory for microbial ecology. ISME J 8:1553–1565
Gogarten JP, Doolittle WF, Lawrence JG (2002) Prokaryotic evolution in light of gene transfer. Mol Biol Evol 19:2226–2238
Goodman SD, Scocca JJ (1988) Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 85:6982–6986
Griffith F (1928) The significance of pneumococcal types. J Hyg (Lond) 27:113–159
Groisman EA, Ochman H (1996) Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791–794
Guglielmini J, De La Cruz F, Rocha EP (2013) Evolution of conjugation and type IV secretion systems. Mol Biol Evol 30:315–331
Hale TL, Keusch GT (1996) Shigella. In: Baron S (ed) Medical microbiology. University of Texas Medical Branch, Galveston, TX
Hanage WP (2013) Fuzzy species revisited. BMC Biol 11:41
Hanage WP (2016) Not so simple after all: bacteria, their population genetics, and recombination. Cold Spring Harb Perspect Biol 8(7):a018069
Hanage WP, Fraser C, Spratt BG (2005) Fuzzy species among recombinogenic bacteria. BMC Biol 3:6
Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H (2001) Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22
Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T (2006) Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007
Hugenholtz P, Skarshewski A, Parks DH (2016) Genome-based microbial taxonomy coming of age. Cold Spring Harb Perspect Biol 8(6):a018085
Jain R, Rivera MC, Lake JA (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A 96:3801–3806
Johnston C, Martin B, Fichant G, Polard P, Claverys JP (2014) Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196
Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, Ding H, Marttinen P, Malmstrom RR, Stocker R, Follows MJ, Stepanauskas R, Chisholm SW (2014) Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344:416–420
Keller I, Wagner CE, Greuter L, Mwaiko S, Selz OM, Sivasundar A, Wittwer S, Seehausen O (2013) Population genomic signatures of divergent adaptation, gene flow and hybrid speciation in the rapid radiation of Lake Victoria cichlid fishes. Mol Ecol 22:2848–2863
Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, Chen F, Lapidus A, Ferriera S, Johnson J, Steglich C, Church GM, Richardson P, Chisholm SW (2007) Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3:e231
Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572
Konstantinidis KT, Rossello-Mora R, Amann R (2017) Uncultivated microbes in need of their own taxonomy. ISME J 11:2399–2406
Krause DJ, Whitaker RJ (2015) Inferring speciation processes from patterns of natural variation in microbial genomes. Syst Biol 64:926–935
Kuo CH, Moran NA, Ochman H (2009) The consequences of genetic drift for bacterial genome complexity. Genome Res 19:1450–1454
Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327
Lang AS, Beatty JT (2007) Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol 15:54–62
Lapierre P, Gogarten JP (2009) Estimating the size of the bacterial pan-genome. Trends Genet 25:107–110
Lapierre M, Blin C, Lambert A, Achaz G, Rocha EP (2016) The impact of selection, gene conversion, and biased sampling on the assessment of microbial demography. Mol Biol Evol 33:1711–1725
Lieberman TD, Flett KB, Yelin I, Martin TR, Mcadam AJ, Priebe GP, Kishony R (2014) Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet 46:82–87
Ludwig W, Klenk HP (2000) Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics. In: Boone DR, Castenholz RW, Garrity GM (eds) Bergey’s manual of systematic biology, 2nd edn. Springer, New York
Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT (2011) Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A 108:7200–7205
Lynch M, Conery JS (2003) The origins of genome complexity. Science 302:1401–1404
Lynch M, Bobay LM, Catania F, Gout JF, Rho M (2011) The repatterning of eukaryotic genomes by random genetic drift. Annu Rev Genomics Hum Genet 12:347–366
Lynch M, Ackerman MS, Gout JF, Long H, Sung W, Thomas WK, Foster PL (2016) Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet 17:704–714
Maddamsetti R, Lenski RE (2018) Analysis of bacterial genomes from an evolution experiment with horizontal gene transfer shows that recombination can sometimes overwhelm selection. PLoS Genet 14:e1007199
Maddamsetti R, Lenski RE, Barrick JE (2015) Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics 200:619–631
Majewski J (2001) Sexual isolation in bacteria. FEMS Microbiol Lett 199:161–169
Majewski J, Cohan FM (1998) The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics 148:13–18
Majewski J, Cohan FM (1999) DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 153:1525–1533
Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG (2000) Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol 182:1016–1023
Mallet J (2007) Hybrid speciation. Nature 446:279–283
Mallet J (2008) Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos Trans R Soc Lond Ser B Biol Sci 363:2971–2986
Mallet J, Beltran M, Neukirchen W, Linares M (2007) Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol Biol 7:28
Mallet J, Besansky N, Hahn MW (2016) How reticulated are species? BioEssays 38:140–149
Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112
Marttinen P, Hanage WP (2017) Speciation trajectories in recombining bacterial species. PLoS Comput Biol 13:e1005640
Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J (2012) Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40:e6
Matic I, Taddei F, Radman M (2000) No genetic barriers between Salmonella enterica serovar typhimurium and Escherichia coli in SOS-induced mismatch repair-deficient cells. J Bacteriol 182:5922–5924
Mayr E (1942) Systematics and the origin of species. Columbia University Press, New-York
Mcinerney JO, Mcnally A, O’connell MJ (2017) Why prokaryotes have pangenomes. Nat Microbiol 2:17040
Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R (2005) The microbial pan-genome. Curr Opin Genet Dev 15:589–594
Mell JC, Shumilina S, Hall IM, Redfield RJ (2011) Transformation of natural genetic variation into Haemophilus influenzae genomes. PLoS Pathog 7:e1002151
Meyer JR, Dobias DT, Medina SJ, Servilio L, Gupta A, Lenski RE (2016) Ecological speciation of bacteriophage lambda in allopatry and sympatry. Science 354:1301–1304
Mira A, Ochman H, Moran NA (2001) Deletional bias and the evolution of bacterial genomes. Trends Genet 17:589–596
Moldovan MA, Gelfand MS (2018) Pangenomic definition of prokaryotic species and the phylogenetic structure of Prochlorococcus spp. Front Microbiol 9:428
Moran NA (1996) Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A 93:2873–2878
Moran NA, Plague GR (2004) Genomic changes following host restriction in bacteria. Curr Opin Genet Dev 14:627–633
Moran NA, Mclaughlin HJ, Sorek R (2009) The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382
Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, Marttinen P (2017) Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol 34:1167–1182
Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS (2016) An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res 26:1612–1625
Novichkov PS, Wolf YI, Dubchak I, Koonin EV (2009) Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol 191:65–73
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Oliveira PH, Touchon M, Rocha EP (2014) The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res 42:10618–10631
Oliveira PH, Touchon M, Rocha EP (2016) Regulation of genetic flux between bacteria by restriction-modification systems. Proc Natl Acad Sci U S A 113:5658–5663
Papke RT, Zhaxybayeva O, Feil EJ, Sommerfeld K, Muise D, Doolittle WF (2007) Searching for species in haloarchaea. Proc Natl Acad Sci U S A 104:14092–14097
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36(10):996–1004
Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT, Beghini F, Malik F, Ramos M, Dowd JB, Huttenhower C, Morgan M, Segata N, Waldron L (2017) Accessible, curated metagenomic data through ExperimentHub. Nat Methods 14:1023–1024
Polz MF, Alm EJ, Hanage WP (2013) Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29:170–175
Popa O, Dagan T (2011) Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol 14:615–623
Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T (2011) Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res 21:599–609
Popa O, Landan G, Dagan T (2017) Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J 11:543–554
Porter SS, Chang PL, Conow CA, Dunham JP, Friesen ML (2017) Association mapping reveals novel serpentine adaptation gene clusters in a population of symbiotic Mesorhizobium. ISME J 11:248–262
Pupo GM, Lan R, Reeves PR (2000) Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A 97:10567–10572
Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ (2009) Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci U S A 106:8605–8610
Retchless AC, Lawrence JG (2007) Temporal fragmentation of speciation in bacteria. Science 317:1093–1096
Retchless AC, Lawrence JG (2010) Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proc Natl Acad Sci U S A 107:11453–11458
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131
Rieseberg LH (1997) Hybrid origins of plant species. Annu Rev Ecol Syst 28:359–389
Riley MA, Lizotte-Waniewski M (2009) Population genomics and the bacterial species concept. Methods Mol Biol 532:367–377
Roberts MS, Cohan FM (1993) The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics 134:401–408
Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A (2009) Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7:828–836
Rolland K, Lambert-Zechovsky N, Picard B, Denamur E (1998) Shigella and enteroinvasive Escherichia coli strains are derived from distinct ancestral strains of E. coli. Microbiology 144(Pt 9):2667–2672
Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, Weinert K, Tenaillon O, Matic I, Denamur E (2009) Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog 5:e1000257
Scocca JJ, Poland RL, Zoon KC (1974) Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J Bacteriol 118:369–373
Seehausen O (2004) Hybridization and adaptive radiation. Trends Ecol Evol 19:198–207
Sela I, Wolf YI, Koonin EV (2016) Theory of prokaryotic genome evolution. Proc Natl Acad Sci U S A 113:11399–11407
Shapiro BJ (2014) Signatures of natural selection and ecological differentiation in microbial genomes. Adv Exp Med Biol 781:339–359
Shapiro BJ, Polz MF (2014) Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol 22:235–247
Shapiro BJ, Polz MF (2015) Microbial speciation. Cold Spring Harb Perspect Biol 7:a018143
Shapiro BJ, David LA, Friedman J, Alm EJ (2009) Looking for Darwin’s footprints in the microbial world. Trends Microbiol 17:196–204
Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G, Polz MF, Alm EJ (2012) Population genomics of early events in the ecological differentiation of bacteria. Science 336:48–51
Shapiro BJ, Leducq JB, Mallet J (2016) What is speciation? PLoS Genet 12:e1005860
Shen P, Huang HV (1986) Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441–457
Sheppard SK, Mccarthy ND, Falush D, Maiden MC (2008) Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237–239
Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, Meric G, Kelly DJ, Cody A, Colles FM, Strachan NJ, Ogden ID, Forbes K, French NP, Carter P, Miller WG, Mccarthy ND, Owen R, Litrup E, Egholm M, Affourtit JP, Bentley SD, Parkhill J, Maiden MC, Falush D (2013) Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol 22:1051–1064
Simmons SL, Dibartolo G, Denef VJ, Goltsman DS, Thelen MP, Banfield JF (2008) Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol 6:e177
Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, De La Cruz F (2010) Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452
Smith JM, Smith NH, O’rourke M, Spratt BG (1993) How clonal are bacteria? Proc Natl Acad Sci U S A 90:4384–4388
Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM (2007) Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318:1449–1452
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849
Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M (2012) Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci U S A 109:18488–18492
Syvanen M (2012) Evolutionary implications of horizontal gene transfer. Annu Rev Genet 46:341–358
Takeuchi N, Cordero OX, Koonin EV, Kaneko K (2015) Gene-specific selective sweeps in bacteria and archaea caused by negative frequency-dependent selection. BMC Biol 13:20
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit Y, Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A 102:13950–13955
Thiergart T, Landan G, Martin WF (2014) Concatenated alignments and the case of the disappearing tree. BMC Evol Biol 14:266
Thingstad T, Lignell R (1997) Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol 13:19–27
Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721
Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, El Karoui M, Frapy E, Garry L, Ghigo J, Gilles A, Johnson J, Le BouguéNec C, Lescat M, Mangenot S, Martinez-JéHanne V, Matic I, Nassif X, Oztas S, Petit M, Pichon C, Rouy Z, Saint Ruf C, Schneider D, Tourret J, Vacherie B, Vallenet D, MéDigue C, Rocha E, Denamur E (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344
Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, Murphy C, Feldgarden M, Wortman J, Clermont D, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P, Rocha EP (2014) The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol 6:2866–2882
Treangen TJ, Rocha EP (2011) Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet 7:e1001284
Trost B, Haakensen M, Pittet V, Ziola B, Kusalik A (2010) Analysis and comparison of the pan-genomic properties of sixteen well-characterized bacterial genera. BMC Microbiol 10:258
Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N (2017) Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27:626–638
Velasco JD (2009) When monophyly is not enough: exclusivity as the key to defining a phylogenetic species concept. Biol Philos 24:473–486
Vernikos G, Medini D, Riley DR, Tettelin H (2015) Ten years of pan-genome analyses. Curr Opin Microbiol 23:148–154
Vos M (2011) A species concept for bacteria based on adaptive divergence. Trends Microbiol 19:1–7
Vos M, Didelot X (2009) A comparison of homologous recombination rates in bacteria and archaea. ISME J 3:199–208
Vulic M, Dionisio F, Taddei F, Radman M (1997) Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci U S A 94:9763–9767
Whitaker RJ, Grogan DW, Taylor JW (2003) Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978
Wiedenbeck J, Cohan FM (2011) Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev 35:957–976
Woese CR, Fox GE (1977) Phylogenetic structure of prokaryotic domain - primary kingdoms. Proc Natl Acad Sci U S A 74:5088–5090
Wright ES, Baum DA (2018) Exclusivity offers a sound yet practical species criterion for bacteria despite abundant gene flow. BMC Genomics 19:724
Yahara K, Didelot X, Ansari MA, Sheppard SK, Falush D (2014) Efficient inference of recombination hot regions in bacterial genomes. Mol Biol Evol 31:1593–1605
Yahara K, Didelot X, Jolley KA, Kobayashi I, Maiden MC, Sheppard SK, Falush D (2015) The landscape of realized homologous recombination in pathogenic bacteria. Mol Biol Evol 33(2):456–471
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zawadzki P, Roberts MS, Cohan FM (1995) The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics 140:917–932
Zhaxybayeva O, Doolittle WF, Papke RT, Gogarten JP (2009) Intertwined evolutionary histories of marine Synechococcus and Prochlorococcus marinus. Genome Biol Evol 1:325–339
Acknowledgments
I thank Bryan McLean, Eduardo PC Rocha, and Kasie Raymann for their opinion on the manuscript. LMB was supported by the National Science Foundation award DEB-1831730.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Bobay, LM. (2020). The Prokaryotic Species Concept and Challenges. In: Tettelin, H., Medini, D. (eds) The Pangenome. Springer, Cham. https://doi.org/10.1007/978-3-030-38281-0_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-38281-0_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-38280-3
Online ISBN: 978-3-030-38281-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)