Abstract
Genome methylation in bacteria is an area of intense interest because it has broad implications for bacteriophage resistance, replication, genomic diversity via replication fidelity, response to stress, gene expression regulation, and virulence. Increasing interest in bacterial DNA modification is coming about with investigation of host/microbe interactions and the microbiome association and coevolution with the host organism. Since the recognition of DNA methylation being important in Escherichia coli and bacteriophage resistance using restriction/modification systems, more than 43,600 restriction enzymes have been cataloged in more than 3600 different bacteria. While DNA sequencing methods have made great advances there is a dearth of method advances to examine these modifications in situ. However, the large increase in whole genome sequences has led to advances in defining the modification status of single genomes as well as mining new restriction enzymes, methyltransferases, and modification motifs. These advances provide the basis for the study of pan-epigenomes, population-scale comparisons among pangenomes to link replication fidelity and methylation status along with mutational analysis of mutLS. Newer DNA sequencing methods that include SMRT and nanopore sequencing will aid the detection of DNA modifications on the ever-increasing whole genome and metagenome sequences that are being produced. As more sequences become available, larger analyses are being done to provide insight into the role and guidance of bacterial DNA modification to bacterial survival and physiology.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Bacterial cellular functions are widely impacted via epigenetic modification, including bacteriophage infection, metabolism, virulence, persistence, replication, and genome plasticity. DNA modification in bacteria is of great interest because it is increasingly being linked to functional regulation processes in the organism and disease progression in mammals (Kumar and Rao 2013). DNA methylation was first recognized in Escherichia coli as part of restriction/modification systems (RMS) that limit and regulate bacteriophage infection. RMS are ubiquitous in the bacterial world with >43,600 RM recognized enzymes in >3600 bacteria (http://rebase.neb.com/rebase/rebase.html) (Roberts et al. 2010). Methylation primarily occurs at N6adenine and C5cytosine in many species, but only N4cytosine is found in bacteria (Wion and Casadesus 2006; Kumar and Rao 2013). Recently, a new modification that regulates the redox status of the cell using DNA modification via a unique multifunctional alteration via phosphothioation was identified (Wang et al. 2019). Subsequently, DNA and RNA methylations were defined to play a central role in bacterial phenotypes that were not encoded in the genome but inherited in bacteria and do regulate gene expression in bacteria. Post-replication modification allows cells to rapidly adjust to local environmental conditions via gene expression changes that are not directly linked to genome variation yet require very dynamic shifts for survival and growth status.
An emerging area of investigation is the role of the microbiome on the host epigenome. Particular interest is paid to the role of the bacterial involvement in host cancer due to dysregulation of gene expression as cancer progresses. A comprehensive review of the state of progress that links infectious agents to cancer and host epigenome proposed that chronic inflammation was involved in the dysregulation of gene expression (Rajagopalan and Jha 2018). An intriguing hypothesis is that bacterial metabolism in utero can have long-lasting effect by regulating epigenetic modification of the maternal and fetal status in utero (Romano and Rey 2018). The complexity of the microbiome composition and metabolism leads one to expect a very complex system for the bacterial community to regulate the host epigenome. Farhana et al. (2018) reviewed the microbiome and its potential role in cancer. Of particular interest is that of Helicobacter pylori since it is associated with multiple states of disease in the progression from normal tissue to cancer with regional and human race differences since it has coevolved with humans for at least 80,000 years (Munoz-Ramirez et al. 2017), and it has a complex lifestyle in the microbial community within a unique location in the body that forces the organism to manage swings in pH, redox, and nutrient sources within minutes.
With the emergence of population genomics and metagenomics and large-scale whole-genome sequencing the vast amount of information has grown rapidly over a short time. With over 350,000 bacterial genomes in the public domain, a new challenge has grown in trying to conduct population epigenomes in bacteria and then associate those changes with change in the host to promote disease. Chen et al. (2014) described a method for population-scale approaches; however, more robust methods are now needed that include metagenome analysis as well.
Comparison of genomes using pangenomes and Big data approaches are progressing to link specific genes and alleles to disease. Population genomics is beginning to emerge (Weis et al. 2016) but it is disconnected to epigenomes and pangenome analysis at this point. Hence, focusing on specific genes and modifications is appropriate and providing results that can be linked to population genomics in the future.
2 Bacterial DNA Modifications and Biological Importance
On a biochemical level, epigenetic modification of the genome changes the accessibility of specific gene clusters and affinity of transcriptional regulators for their cognate promoters. This modulation of transcription accessibility and promoter affinity in turn translates to changes in bacterial response to environmental stimuli. Because epigenetic modifiers, such as RM systems and specific methyltransferases (MTases) themselves, are encoded on the chromosome as well as on plasmids, these elements can be transmitted vertically as a result of replication as well as horizontally as a result of horizontal gene transfer either via conjugation or phage. As mentioned above, DNA modification systems serve to identify and eliminate foreign DNA, but these DNA modifications also serve important roles in cell cycle progression, DNA repair, and regulation of gene expression.
2.1 Bacterial Histone-Like Proteins
Like eukaryotic histones, bacterial histone-like proteins assist in compacting the chromosome into a nucleoid structure (Thanbichler et al. 2005). Histone-like proteins can be classified into four different categories: histone-like proteins (HU), histone-like nucleoid structuring proteins (H-NS), integration host factors (IHF), and factors for inversion stimulation (FIS), further reviewed in Dorman and Deighan (2003) and Anuchin et al. (2011). To accomplish this task, bacteria utilize histone-like proteins to organize their DNA to minimize space utilization but also to regulate the expression of their DNA. These proteins work in a concerted manner to bind DNA and facilitate supercoiling into a nucleoid structure and regulate gene expression, these mechanisms were extensively reviewed previously (Dorman and Deighan 2003; Thanbichler et al. 2005; Dorman 2013; Takahashi 2014; Grainger 2016). Throughout the cell cycle, different histone-like proteins peak in concentration to regulate genes sets responsible for the progression of an actively replicating cell to a stationary phase cell, indicating that each one plays a unique role during specific stages of growth. Cycling histone-like proteins indicates that the pan-epigenome changes at different phases of growth. In addition to being related to different growth phases, expression of specific histone-like proteins is also induced in response to environmental stresses. The ability of environmental stimuli to change histone association with DNA suggests that pan-epigenetic shifts occur when an organism adapts to its environment. Examples are evident in the existence of microbes adapted to live in extreme environments as well as pathogens, such as Brucella, that are specifically adapted to live in their host. While these microbes no longer possess genes found in related species, it was epigenetic selection that led to the refinement of these genomes. Sustained pan-epigenetic shifts result in perpetually inactivated genes that are subsequently lost in future generations, resulting in differentiation between DNA modification and genotypes.
Although DNA methylation is frequently associated with RM systems and bacterial “immunity” against sources of foreign DNA, we are just beginning to understand the global impacts of DNA methylation on transcriptional regulation of gene expression. In addition to protein–DNA interactions affected by methylation, DNA modifications also regulate bacterial histone-like protein binding to DNA.
While MTases may indirectly impact gene expression through modulating histone-like protein–DNA interactions, MTases directly influence gene expression through the presence of recognition motifs located in promoter regions and protein-binding sites of genes. The methylation state of these regions work by modulating the affinity of RNA polymerase and transcriptional regulators such as leucine-responsive repeat protein (Lrp) and catabolite activator protein (Cap) to specific genes, among which include dnaA, ppiA, yhiP, and the pap operon (Tavazoie and Church 1998; Marinus and Casadesus 2009).
RM systems play a major role in bacterial immunity against foreign DNA. Another component of the bacterial “immune system” was recently discovered, termed clustered regularly interspaced palindromic repeats/CRISPR-associated (CRISPR/Cas). CRISPR systems are detectable in 1126 of the 2480 genomes analyzed to date (Grissa et al. 2007). Similar to phase variable regions of the genome, CRISPR/Cas systems are composed of short, conserved, DNA repeat sequences interspersed by stretches of variable sequences with cas genes adjacent to these regions. CRISPR/Cas systems recognize foreign nucleic acids, targeting them for degradation via RNA interference effector complexes composed of Cas proteins and CRISPR RNAs (Gasiunas et al. 2013). Though no associations between MTases and CRISPR/Cas have been proven, Hernández-Lucas et al. determined that Salmonella Typhi casA is under H-NS and Lrp regulation (Medina-Aparicio et al. 2011). In addition to immunity, CRISPR/Cas systems are also hypothesized to affect DNA mismatch repair with E. coli Cas1 involved in DNA segregation and mismatch repair (Babu et al. 2011; Westra et al. 2012). MTases and CRISPRs both share a number of common interacting partners involved in transcriptional regulation including Lrp and H-NS. While much remains to be learned about additional cellular roles of these systems, it is not improbable to expect a synergistic interaction in orchestrating essential cell processes.
2.2 DNA Modifications
Bacteria encode numerous restriction-modification (RM) systems that can be categorized into four main types. RM systems include the restriction endonuclease (REase), methyltransferase (MTase), and the specificity protein which facilitate targeted RM enzymatic activity to specific regions of DNA. RM systems require a specific unit, which enables RM targeting to a DNA recognition domain, a methyltransferase that modifies DNA with a methyl group, and an endonuclease that cleaves DNA (REase) with four types of RM systems described to date and catalogued in Rebase (Roberts et al. 2010). Briefly, Type I is characterized by an oligomeric MTase and REase complex with restriction occurring at variable distances from the recognition site. As the largest category with over 16,000 MTases identified, Type II system fall into numerous subcategories and are composed of either discreet or fused, MTase and REase subunits that cleave at or near the recognition site. Type III system cut at a fixed site away from the recognition sequence with the restriction enzyme activity contingent on association with the cognate MTase. Like Type I, Type IV system cleave at a variable distance from the recognition site but unlike the other three systems, the Type IV system is able to recognize and cleave hydroxymethylated and phosphorothioated DNA in addition to methylated DNA (Vasu and Nagaraja 2013; Loenen et al. 2014).
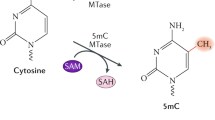
Originally discovered as a protective mechanism against bacteriophage infection, MTases selectively transfer the methyl group from SAM to the nitrogen atoms at position 4 in cytosine and position 6 of adenine (m4C, m6A) or the fifth carbon of cytosine (m5C) within specific sequence motifs along the bacterial genome identified by the RM system recognition domain (Wilson 1991). These methylated sequences are resistant to endonuclease digestion by the restriction enzyme and are recognized by the RM system as a means of establishing self from nonself. Any phage DNA entering the host is assessed by the RM system and digested by the RM endonuclease if methylation is not detected by the corresponding recognition domain. To circumvent host restriction of phage DNA, bacteriophage often introduces their own MTases during infection. Due to the nature of RM enzyme–DNA dynamics, these MTases are often retained by the host following bacteriophage infection and transferred to subsequent generations, giving rise to orphan MTases lacking a reciprocal restriction enzyme (Labrie et al. 2010; Murphy et al. 2013).
Early experiments involving manipulation of RM systems produced viable cells with r + m + and r-m + phenotypes. Interestingly, an r + m- phenotype was lethal, suggesting that in the absence of DNA methylation, restriction enzymes will digest self-DNA, resulting in cell death (Arber 1965). In studying postsegregational killing by RM systems, Kobayashi et al. observed a larger amount of MTases molecules relative to REase in steady-state cells. However, dysregulation of cellular MTase and REase levels led to increased cell death due to Res-induced double-strand breaks in the chromosome (Ichige and Kobayashi 2005). These results further highlight a characteristic true of all RM systems in which MTases are fully functional without the cognate restriction enzyme; however, the restriction enzyme activity is contingent on the presence of the MTase. Easy acquisition and retention of foreign MTases—termed orphan MTases—by host bacteria contributes to the increased diversity of MTases in relation to restriction enzymes with possible methyltransferase sources being mobile elements acquired through transduction or mating events (Murphy et al. 2013).
DNA Adenine Methylation DAM
DNA adenine methylation (Dam) is the predominant methylation found in bacteria and is accomplished by bacterial methyltransferases (MTases). Dam MTases are widespread throughout all genera of bacteria, with some MTases sharing the same recognition motif and other MTase recognition sites being species, if not strain, specific. The presence of hydrophobic methyl groups either on both strands of DNA (fully methylated) or a single strand of DNA (hemi methylated) serve to modulate gene expression by way of modulating the affinity of DNA-binding proteins for specific regions of DNA.
Survival in a niche environment such as the human body requires careful and concerted regulation of numerous genes, ranging from stress response and nutrient acquisition to manipulation of host processes in the case of pathogenic bacteria. Although bacterial pathogens have coevolved with their hosts (Hongoh et al. 2005), the standard transmission cycle of some pathogens dictate that they may spend some time outside of their human host and in environments that are suboptimal in moisture and nutrients but can contain antimicrobial compounds (Harb et al. 2000). Transitioning from an environmental lifestyle to a host-adapted lifestyle requires a large shift in the gene expression and protein profile of a pathogen. With the magnitude of gene regulation needed to facilitate this lifestyle change, it is reasonable to consider the role of epigenetics in driving these changes (Low et al. 2001).
E. coli
The pap operon of E. coli encodes the pyelonephritis-associated pilus. While pap is under methylation-mediated transcriptional control, Pap expression is also regulated by methylation-mediated phase variation. Mechanistically, Dam competes with transcriptional regulators, such as Lrp, a global transcriptional activator, for access to recognition domains wherein methylation of the domain determines the pilus ON/OFF state (Casadesus and Low 2006). Similar mechanisms governing pilus formation and phase variation are also documented in many other bacteria including Salmonella, S. aureus, H. influenza, Neisseria, and H. pylori (Srikhanta et al. 2005, 2011).
Salmonella
This organism is broadly modified (Table 1) over the genome with specific motifs. Within the same Salmonella virulence plasmid, H-NS represses finP in a Dam-dependent manner while repressing traJ in a Dam-independent manner. These observations bring to light the impact of structural differences in nucleoids of dam + vs dam- genomes and the outcome of these structural differences on gene expression (Marinus and Casadesus 2009). In addition to histone-like proteins, DNA methylation, specifically adenine methylation (Dam) is known to be involved in regulating host colonization. PhoP, a master regulator of Salmonella virulence, binds DNA in a dam-dependent manner (Heithoff et al. 1999). Deletion or over expression of an MTase results in whole genome-wide change in transcription profiles. While Salmonella Typhimurium Dam mutants do not exhibit growth-related deficiencies, Dam-deficient Salmonella exhibits a 10,000-fold increase in the lethal dose required to kill 50% of a mouse population (LD50) (Low et al. 2001). Transcriptional profiling of Dam-deficient Salmonella attributes attenuation to an induction of spvB, along with over 35 other infection-associated genes and a reduction in sipABC transcripts (Garcia-Del Portillo et al. 1999).
The amount of information in specific organisms that have a minor role in disease or lack a large amount of whole genome sequence has very little pan-epigenome information. Chen et al. (2017) examined the epigenome of L. monocytogenes (Table 2) to find a complex pattern of modification that was not observed to be associated with pathogenicity. Virulence genes were heavily methylated, but no observable pattern emerged to uncover how methylation was involved in virulence.
DNA Cytosine Methylation (DCM)
Unlike adenine methylation that has been functionally characterized in numerous bacterial systems, DNA cytosine methylation (Dcm) remains relatively understudied. Best characterized in E. coli, Dcm appears to confer resistance against restriction by the REase, EcoRII (Bigger et al. 1973; Boye and Lobner-Olesen 1990). Functionally, Dcm acts as an antitoxin against EcoRII restriction. Because Dcm and EcoRII share the same recognition sequence—CmCWGG—Dcm is able to methylate sites that would otherwise be targeted for EcorII restriction (Palmer and Marinus 1994). In this manner, Dcm serves a protective function against a parasitic RM system (Takahashi et al. 2002). Dcm is also associated with mobile element rearrangements in the E. coli genome involving bacteriophage lambda recombination and TN3 transposition (Korba and Hays 1982; Yang et al. 1989). On a whole genome level, evidence suggests that Dcm is involved in transcriptional and translational regulation of ribosome activity to decrease the expression of ribosomal proteins during stationary phase (Militello et al. 2012).
Phosphorothioate Modification
A third, recently discovered DNA modification that naturally occurs in bacteria is phosphorothioate (PT) modification wherein the oxygen atom in a phosphate moiety of the DNA backbone is replaced by sulfur (Eckstein 2014). The ability to carry out PT modifications is contingent on the presence of the dnd gene clusters, dndABCDE, the modification component, and dndFGH, the restriction component although their presence can be mutually exclusive (Tong et al. 2018). First discovered in Streptomyces lividans, informatics analyses of dnd gene clusters has since revealed a wide distribution of PT modifications in bacterial genomes (He et al. 2007; Wang et al. 2011, 2019). Abrogation of PT modifications led to increased double-stranded DNA breaks in Salmonella and oxidative stress due to significant metabolic changes in Pseudomonas fluorescens (Cao et al. 2014; Gan et al. 2014; Tong et al. 2018).
Undiscovered Modifications
Next-generation sequencing techniques that incorporate measurement of polymerase kinetics can detect structural differences to individual nucleotides that would otherwise have been overlooked (Rhoads and Au 2015). By comparing the pattern of polymerase kinetics to previously characterized patterns, we can informatically identify DNA modifications at the single nucleotide level and characterize epigenetic patterns on the whole genome level (Schadt et al. 2013). The use of this technology in whole genome sequencing has also recorded polymerase kinetics patterns that are not yet associated with a known DNA modification (Chen et al. 2017). These data suggest that there is unprecedented diversity to epigenetic modifications that we have yet to uncover. Epigenetic modifications that have been characterized thus far are responsible for numerous physiological processes including defense against foreign DNA, gene regulation, and DNA replication and mismatch repair. The implications of uncharacterized modifications on epigenomic regulation potentially have far-reaching implications for interactions within a niche and interaction with the host for survival and persistence. As additional advances are made in next-generation sequencing and RNAseq, it may be possible to define methylation directly in situ, which is a current limitation.
2.3 DNA Replication and Chromosome Sorting
Bacteria encode proteins near their chromosomal origin of replication (oriC) that facilitate the timing of replication initiation and help to carry out the chromosome segregation during replication (Ogden et al. 1988; Boye and Lobner-Olesen 1990; Campbell and Kleckner 1990). Due to the time-sensitive nature of replication initiation, DNA replication-associated protein levels must be tightly coordinated with cellular replicative machinery. To accomplish this task, bacteria encode a higher density of GATC methylation sites around the origin of replication and utilize DNA methylation to modulate the affinity of replication-associated proteins to DNA. Methylation around oriC regulates the recruitment of replication initiation proteins including the initiator of replication, DnaA. Furthermore, GATC methylation motifs also exist in the promoter region of dnaA, allowing for transcriptional regulation of replication (Campbell and Kleckner 1990). During DNA replication, both copies of the chromosome must be accurately sorted into the corresponding cell. After replication, DNA is in a hemi-methylated state. Methylation at oriC sequesters the origin replication initiation and prevents reinitiation of DNA replication. Additionally, global hemi-methylation of newly replicated DNA facilitates chromosome binding to designated areas of the cell membrane such that individual chromosomes may be accurately partitioned into each daughter cell (Ogden et al. 1988).
2.4 Mismatch Repair and Evolution
Bacterial DNA polymerases are capable to replicating DNA with high fidelity, but replication errors still arise at a rate of 10−9 to 10−11 errors per base pair (Drake et al. 1998). When these replication errors arise, the cell must have a way of identifying the correct template with which to correct the mistake. Template and newly replicated strands of DNA are differentially methylated to differentiate from one another with the template being methylated and the newly replicated strand remaining unmethylated. First described in Streptococcus pneumoniae and further characterized in E. coli, this methyl-directed mismatch repair system was identified as MutHLS (Glickman and Radman 1980; Claverys and Lacks 1986) (Fig. 1). MutS binds to mismatched base pairs while the methyl-sensitive endonuclease MutH nicks the DNA at the mismatched site. MutL recruits the DNA repair machinery to correct the mismatch. Both the loss of MTases and overexpression of MTases are correlated with deficient mismatch repair due to a dysregulation between methylation and DNA replication kinetics. In dam mutants, the inability to methylate the template strand leads to inaccurate mismatch repair and vertical transmission of mutations arising from DNA replication. Dam mutants are unable to methylate the template strands of replicated DNA, leading MutHLS inability to identify the strand of DNA containing the mutation for mismatch repair. In this regard, the pan-epigenome directly influences the accumulation of SNPs that arise during replication. Due to the mobile nature of RMS systems, over time the loss or acquisition of additional MTase systems may influence the global methylation status of a genome.
3 Epigenetic Detection Methods and Approaches
Nucleotide modification by methylation is a prevalent feature in living organisms. In bacteria, base methylation is a form of defense system against bacteriophage or foreign genetic material. The defense system works by detecting sequence motifs of nucleotides and cuts it using an endonuclease as a preemptive strike against foreign genome. Bacterial DNA is spared from the cutting with the action of the methylase. This is known as the restriction-modification system (RMS). Aside from defensive function, the restriction modification system also performs genomic regulatory functions in bacteria. Due to the huge impact of the restriction modification system in the lifestyle of bacteria with regard to pathogenicity, prokaryotic epigenomics is an emerging field primarily driven by recent technological advancement in sequencing capability. The transformational aspect is mainly on the scalability of methylation analysis at the genomic level. This has opened up doors for genome-wide methylation analysis.
What are the key considerations in doing large-scale high-throughput epigenomics research? Genome-wide methylation projects’ considerations are determined by costs, ease of library construction and preparation, access to equipment or core facility, availability of suitable kits for library construction and downstream bioinformatic analysis. The level of resolution of epigenomic modification data from crude to precise distinguishes the possible technological options appropriate for the pipeline. The above-mentioned considerations as well as the underlying technology will be covered in the succeeding sections.
3.1 Pre-sequencing Methods for Genome Methylation: LC-MS, HPLC-UV, and ELISA
The pre-sequencing methods are generally used for basic research and their capability to quantify methylation at the genomic scale. While this ability to quantify methylation at the genome scale provides a big picture setting of methylation, mapping the methylation sites to the specific regions in the genome is not possible. The scalability for population-scale bacterial epigenomics is limited and hence has limited the applicability of these methods to a few niche research papers.
The key steps in the analytical workflows are DNA extraction, genomic fragmentation, enrichment, and quantification using chromatography or mass spectrometry. The options for genomic fragmentation are thermal, chemical, and enzymatic hydrolysis. The resulting digested DNA monomers is enriched using size-exclusion, liquid extraction, solid phase extraction, or preparative liquid chromatography. Analyte ions are separated by the mass-to-charge ratios in mass spectrometry, allowing binning of the DNA monomers (Tretyakova et al. 2013).
Genome wide methylation using analytical methods particularly HPLC-based methods have been recently described (Yotani et al. 2018). High-performance liquid chromatography-ultraviolet (HPLC-UV) enables quantification and identification by separating the different components. This is accomplished by pushing the components using pressurized liquid solvent through a column filled with solid adsorbent material. The differences between the materials result to variation in flow rates allowing separation of the components. In bacterial DNA methylation analysis, this method is applied to quantify the separated methylated and unmethylated deoxynucleosides.
For crude global methylation analysis, numerous commercial ELISA (enzyme-linked immunosorbent assay) kits are available. The high level of variance is the primary reason for the lack of precision of ELISA kits in epigenomics, but the ease of use is sufficient to capture huge differences in methylation. The target DNA is immobilized on ELISA plate and specific primary antibody against methylated nucleoside is applied followed by a secondary antibody that can be detected using colorimetric methods.
The requirement for specialized equipment for LC-MS and HPLV-UV has restricted the use of the following methods for genome-wide methylation. While relative quantification is possible, mapping the methylation is not possible and hence population-scale analysis is not possible. The technical challenges of doing the work hinders its large-scale application.
3.2 Next-Generation Sequencing-Based Methods
The key shortcoming in using analytical methods for bacterial epigenomics is inability to identify methylation loci. This deficiency has predominantly filled by next-generation sequencing technology that can simultaneously capture sequence and methylation data (Fig. 2). The prevailing choice for combined sequencing and methylation platform is single molecule real-time (SMRT) sequencing by PacBio. Data is captured for 6mA, 4mC, and 5mC parallel to sequencing data based on the kinetics of DNA synthesis reactions. This enables genome-wide mapping of methylated and unmethylated loci. Modified bases have not been a routinely included in the Sanger-based sequence analysis and has posed significant technological challenge until the arrival next-generation sequencing options. DNA treatment with bisulfite converts unmodified cytosine to uracil, enabling discrimination between modified and unmodified cytosine using various sequencing platform.
SMRT sequencing follows the typical workflow for next-generation sequencing with library construction after DNA extraction (Kong et al. 2017). The protocols for automated PacBio 10 kb library construction have been published, which can immensely improve efficiency of performing epigenomic research. A crucial requirement for successful high-throughput sequencing run is high molecular weight genomic DNA. Agilent 2200 TapeStation Nucleic Acid System has been used to determine the quantity and size distribution of purified genomic DNA (Kong et al. 2014) as well as the 260/280 and 260/230 ratio using Nanodrop 2000 UV–vis spectrophotometer (ThermoFisher Scientific, Waltham MA). The DNA integrity number (DIN) is a suitable tool for determining the quality of genomic DNA for further processing (Kong et al. 2016) and methods exist for automated construction of the sequencing library (Kong et al. 2017). The core basis for SMRT sequencing is based on restrictions of light illumination of immobilized target DNA and polymerase using zero-mode waveguide (Rhoads and Au 2015). Signal detection of the cleaved fluorescent dye from the nucleotide molecule is the basis for base calling. The bulk of the most technically challenging aspect of the analysis is within the post sequencing bioinformatic pipeline. DNA methylation detection and quantification analysis are done in PacBio SMRT analysis platform (http://www.pacb.com/devnet/code.html). After sequencing, raw reads are trimmed to remove adapter sequences and then aligned to a reference using BLASR (v1) (Chaisson and Tesler 2012). DNA methylated sites are then determined using kinetic analysis of the genomic alignment. MotifFinder clusters the methylated sites to motifs targeted by methylases. This platform also allows discovery of novel restriction-modification genes. Homology is inferred bioinformatically using databases like SeqWare for cloud applications (O’Connor et al. 2010).
The development in sequencing technology allowed large-scale analysis of prokaryotes (Blow et al. 2016). Base resolution methylation was captured in unprecedented detail and scale using SMRT sequencing initially. The variety of methylation was found on about 800 different loci in this study, indicative of precise specificities of methylation present in the bacterial organism. With the use of SMRT sequencing, the methylation repertoire was significantly increased. This highlights the key advantage of SMRT sequencing to further enhanced the recognition specificities of the methylase. Novel mechanistic epigenomic findings include: Type I RM system cleavage of DNA at large distances from their recognition sites, while both Type II and Type III systems incomplete cleavage pattern. This epigenomic feature is problematic for digestion-based analytical methods. The predilection of these RMS is toward m4C and m6A, which are readily detected by SMRT sequencing. Another understudied aspect of methylation is the orphaned methylases, which are common in prokaryotes. This relatively understudied group includes 100 Type II methylases. One novel discovery is potential regulatory control due to the genomic pattern associated with the orphan methylases which are located on noncoding sequences upstream of genes. This potential regulatory role was is widely distributed across the prokaryotic organism. In another study, a deeper resolution analysis such as identification and quantification of methylation motifs, correlation with methylases of methylation motifs using REBASE (Roberts et al. 2015) and identification of orphaned methylases has been done in large scale in organisms like Listeria (Chen et al. 2017). This study reported lineage- and clade-specific patterns of restriction-modification system (RMS). Type II RMS dominates with its presence in 256 out of 302 genomes, followed by Type I with 110 genomes, Type IV with 73 and lastly by Type III with 25 genomes. Methylation motifs were also described. These studies highlight the large-scale applicability of sequencing-based epigenomic study to unravel population-scale dynamics and patterns.
On a mechanistic level using fine-scale analysis, Fang et al. explored 6 mA methylation in a Shiga toxin-producing a strain of E. coli 0104:H4 Germany outbreak isolate predicted to produce 10 methylases that result in the 6-mA modification (Fang et al. 2012). A phage-encoded modification system capable of targeting hundreds of loci within the E. coli 0104:H4 isolate. This discovery of phage-encoded modification system-associated virulence had no prior examples in E. coli, illustrating the immense power to untangle epigenomic clues using sequencing platforms.
4 Conclusion and Future Direction
The epigenomic studies relied heavily on bioinformatics to deduce motifs that were highly enriched by modification with specific methylases. These studies discovered novel methylase specificities, quantified methylation activity, identified novel enzyme activity, which targets only one strand of DNA and promiscuous gene lacking specificity. Such precision is only possible with sequencing technology coupled with methylation detection capability. As sequencing technologies advance, the definition of modification will become increasingly important in biological function interpretation. A current limitation is that the vast amount of whole genome sequence and the limited number of methods to locate and estimate the modifications. A proxy for this limitation is to examine the RMS enzymes, which is interesting, but not direct enough to derive biologically accurate information. This method also suffers from informatics methods that can be applied on a comparative population scale, as can be done with pangenomes, but not pan-methylomes for bacteria. MethBank is available for a few mammals and plants (Li et al. 2018). The rate of bacterial genome production is only increasing. As such, a need exists to interrogate methylome of the organism at the speed of sequencing. This is not available and is a severe limitation in understanding bacterial growth, survival, and association; which is also true of metagenome interrogation as well. A great step forward would be to have a similar database for bacteria with the ability to allow pangenome and pan-methylome comparisons.
The field is poised to link the bacterial methylation status with the host methylation composition as it relates to disease. However, the dynamic nature of the microbiome, gene expression, and methylation in the bacterial component is a substantial challenge. Initial stages of examining the microbiome sequence for RMS enzymes are a starting point that will aid in understanding the complement of modifications that are possible. The beginning of this work has started in cancer progression and to some degree single organisms, such as H. pylori, in the development of various stages of cancer progression.
Bacterial metagenome production will increase with the expanded use of real-time sequencing technologies, such as nanopores. However, limitations in analysis and the dynamic nature of the bacterial DNA modification must be addressed to make substantial progress in linking it to phenotype. Future prospects of examining methylation are very exciting and there are many needs in the bioinformatic comparative analysis, especially in pathogens associated with chronic diseases.
References
Anuchin AM, Goncharenko AV, Demidenok OI, Kaprel'iants AS (2011) Histone-like proteins of bacteria (review). Prikl Biokhim Mikrobiol 47(6):635–641
Arber W (1965) Host-controlled modification of bacteriophage. Annu Rev Microbiol 19:365–378
Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, Phanse S, Joachimiak A, Koonin EV, Savchenko A, Emili A, Greenblatt J, Edwards AM, Yakunin AF (2011) A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol 79(2):484–502
Bigger CH, Murray K, Murray NE (1973) Recognition sequence of a restriction enzyme. Nat New Biol 244(131):7–10
Blow MJ, Clark TA, Daum CG, Deutschbauer AM, Fomenkov A, Fries R, Froula J, Kang DD, Malmstrom RR, Morgan RD, Posfai J, Singh K, Visel A, Wetmore K, Zhao Z, Rubin EM, Korlach J, Pennacchio LA, Roberts RJ (2016) The Epigenomic landscape of prokaryotes. PLoS Genet 12(2):e1005854
Boye E, Lobner-Olesen A (1990) The role of dam methyltransferase in the control of DNA replication in E. coli. Cell 62(5):981–989
Campbell JL, Kleckner N (1990) E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62(5):967–979
Cao B, Cheng Q, Gu C, Yao F, DeMott MS, Zheng X, Deng Z, Dedon PC, You D (2014) Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate-based restriction system in salmonella. Mol Microbiol 93(4):776–785
Casadesus J, Low D (2006) Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70(3):830–856
Chaisson MJ, Tesler G (2012) Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238
Chen P, Jeannotte R, Weimer BC (2014) Exploring bacterial epigenomics in the next-generation sequencing era: a new approach for an emerging frontier. Trends Microbiol 22(5):292–300
Chen P, den Bakker HC, Korlach J, Kong N, Storey DB, Paxinos EE, Ashby M, Clark T, Luong K, Wiedmann M, Weimer BC (2017) Comparative genomics reveals the diversity of restriction-modification systems and DNA methylation sites in Listeria monocytogenes. Appl Environ Microbiol 83(3)
Claverys JP, Lacks SA (1986) Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev 50(2):133–165
Dorman CJ (2013) Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem Soc Trans 41(2):542–547
Dorman CJ, Deighan P (2003) Regulation of gene expression by histone-like proteins in bacteria. Curr Opin Genet Dev 13(2):179–184
Drake JW, Charlesworth B, Charlesworth D, Crow JF (1998) Rates of spontaneous mutation. Genetics 148(4):1667–1686
Eckstein F (2014) Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther 24(6):374–387
Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, Deikus G, Clark TA, Luong K, Murray IA, Davis BM, Keren-Paz A, Chess A, Roberts RJ, Korlach J, Turner SW, Kumar V, Waldor MK, Schadt EE (2012) Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30(12):1232–1239
Farhana L, Banerjee HN, Verma M, Majumdar APN (2018) Role of microbiome in carcinogenesis process and epigenetic regulation of colorectal cancer. Methods Mol Biol 1856:35–55
Gan R, Wu X, He W, Liu Z, Wu S, Chen C, Chen S, Xiang Q, Deng Z, Liang D, Chen S, Wang L (2014) DNA phosphorothioate modifications influence the global transcriptional response and protect DNA from double-stranded breaks. Sci Rep 4:6642
Garcia-Del Portillo F, Pucciarelli MG, Casadesus J (1999) DNA adenine methylase mutants of salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc Natl Acad Sci USA 96(20):11578–11583
Gasiunas G, Sinkunas T, Siksnys V (2013) Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci 71(3):449–465
Glickman BW, Radman M (1980) Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci USA 77(2):1063–1067
Grainger DC (2016) Structure and function of bacterial H-NS protein. Biochem Soc Trans 44(6):1561–1569
Grissa I, Vergnaud G, Pourcel C (2007) The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172
Harb OS, Gao LY, Abu Kwaik Y (2000) From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ Microbiol 2(3):251–265
He X, Ou HY, Yu Q, Zhou X, Wu J, Liang J, Zhang W, Rajakumar K, Deng Z (2007) Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol 65(4):1034–1048
Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ (1999) An essential role for DNA adenine methylation in bacterial virulence. Science 284(5416):967–970
Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Vongkaluang C, Noparatnaraporn N, Kudo T (2005) Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol 71(11):6590–6599
Ichige A, Kobayashi I (2005) Stability of EcoRI restriction-modification enzymes in vivo differentiates the EcoRI restriction-modification system from other postsegregational cell killing systems. J Bacteriol 187(19):6612–6621
Kong N, Ng W, Azarene F, Carol Huang B, Kelly L, Weimer BC (2014) Quality control of library construction pipeline for PacBio SMRTbell 10kb library using Agilent 2200 TapeStation. Agilent Technologies, Santa Clara, CA. https://doi.org/10.13140/RG.2.1.4339.4644
Kong N, Ng W, Cai L, Leonardo A, Weimer BC (2016) Integrating the DNA integrity number (DIN) to assess genomic DNA (gDNA) quality control using the Agilent 2200 TapeStation system. Agilent Technologies, Santa, Clara, CA, pp 1–6
Kong N, Ng W, Thao K, Agulto R, Weis A, Kim KS, Korlach J, Hickey L, Kelly L, Lappin S, Weimer BC (2017) Automation of PacBio SMRTbell NGS library preparation for bacterial genome sequencing. Stand Genomic Sci 12:27
Korba BE, Hays JB (1982) Partially deficient methylation of cytosine in DNA at CCATGG sites stimulates genetic recombination of bacteriophage lambda. Cell 28(3):531–541
Kumar R, Rao DN (2013) Role of DNA methyltransferases in epigenetic regulation in bacteria. In: Kundu TK (ed) Epigenetics: development and disease. Springer, Dordrecht, pp 81–102
Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8(5):317–327
Li R, Liang F, Li M, Zou D, Sun S, Zhao Y, Zhao W, Bao Y, Xiao J, Zhang Z (2018) MethBank 3.0: a database of DNA methylomes across a variety of species. Nucleic Acids Res 46(D1):D288–D295
Loenen WA, Dryden DT, Raleigh EA, Wilson GG, Murray NE (2014) Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res 42(1):3–19
Low DA, Weyand NJ, Mahan MJ (2001) Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun 69(12):7197–7204
Marinus MG, Casadesus J (2009) Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev 33(3):488–503
Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernandez AL, Vazquez A, Olvera L, Gutierrez-Rios RM, Calva E, Hernandez-Lucas I (2011) The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol 193(10):2396–2407
Militello KT, Simon RD, Qureshi M, Maines R, VanHorne ML, Hennick SM, Jayakar SK, Pounder S (2012) Conservation of Dcm-mediated cytosine DNA methylation in Escherichia coli. FEMS Microbiol Lett 328(1):78–85
Munoz-Ramirez ZY, Mendez-Tenorio A, Kato I, Bravo MM, Rizzato C, Thorell K, Torres R, Aviles-Jimenez F, Camorlinga M, Canzian F, Torres J (2017) Whole genome sequence and phylogenetic analysis show helicobacter pylori strains from Latin America have followed a unique evolution pathway. Front Cell Infect Microbiol 7:50
Murphy J, Mahony J, Ainsworth S, Nauta A, van Sinderen D (2013) Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol 79(24):7547–7555
O’Connor BD, Merriman B, Nelson SF (2010) SeqWare query engine: storing and searching sequence data in the cloud. BMC Bioinformatics 11(Suppl 12):S2
Ogden GB, Pratt MJ, Schaechter M (1988) The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell 54(1):127–135
Palmer BR, Marinus MG (1994) The dam and dcm strains of Escherichia coli—a review. Gene 143(1):1–12
Rajagopalan D, Jha S (2018) An epi(c)genetic war: pathogens, cancer and human genome. Biochim Biophys Acta Rev Cancer 1869(2):333–345
Rhoads A, Au KF (2015) PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13(5):278–289
Roberts RJ, Vincze T, Posfai J, Macelis D (2010) REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 38(Database issue):D234–D236
Roberts RJ, Vincze T, Posfai J, Macelis D (2015) REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43(Database issue):D298–D299
Romano KA, Rey FE (2018) Is maternal microbial metabolism an early-life determinant of health? Lab Anim (NY) 47(9):239–243
Schadt EE, Banerjee O, Fang G, Feng Z, Wong WH, Zhang X, Kislyuk A, Clark TA, Luong K, Keren-Paz A, Chess A, Kumar V, Chen-Plotkin A, Sondheimer N, Korlach J, Kasarskis A (2013) Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res 23(1):129–141
Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP (2005) The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci USA 102(15):5547–5551
Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, Robins-Browne RM, Jennings MP (2011) Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One 6(12):e27569
Takahashi K (2014) Influence of bacteria on epigenetic gene control. Cell Mol Life Sci 71(6):1045–1054
Takahashi N, Naito Y, Handa N, Kobayashi I (2002) A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J Bacteriol 184(22):6100–6108
Tavazoie S, Church GM (1998) Quantitative whole-genome analysis of DNA-protein interactions by in vivo methylase protection in E. coli. Nat Biotechnol 16(6):566–571
Thanbichler M, Wang SC, Shapiro L (2005) The bacterial nucleoid: a highly organized and dynamic structure. J Cell Biochem 96(3):506–521
Tong T, Chen S, Wang L, Tang Y, Ryu JY, Jiang S, Wu X, Chen C, Luo J, Deng Z, Li Z, Lee SY, Chen S (2018) Occurrence, evolution, and functions of DNA phosphorothioate epigenetics in bacteria. Proc Natl Acad Sci USA 115(13):E2988–E2996
Tretyakova N, Villalta PW, Kotapati S (2013) Mass spectrometry of structurally modified DNA. Chem Rev 113(4):2395–2436
Vasu K, Nagaraja V (2013) Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77(1):53–72
Wang L, Chen S, Vergin KL, Giovannoni SJ, Chan SW, DeMott MS, Taghizadeh K, Cordero OX, Cutler M, Timberlake S, Alm EJ, Polz MF, Pinhassi J, Deng Z, Dedon PC (2011) DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci USA 108(7):2963–2968
Wang L, Jiang S, Deng Z, Dedon PC, Chen S (2019) DNA phosphorothioate modification-a new multi-functional epigenetic system in bacteria. FEMS Microbiol Rev 43(2):109–122
Weis AM, Storey DB, Taff CC, Townsend AK, Huang BC, Kong NT, Clothier KA, Spinner A, Byrne BA, Weimer BC (2016) Genomic comparisons and zoonotic potential of campylobacter between birds, primates, and livestock. Appl Environ Microbiol 82:7165–7175
Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J (2012) The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet 46:311–339
Wilson GG (1991) Organization of restriction-modification systems. Nucleic Acids Res 19(10):2539–2566
Wion D, Casadesus J (2006) N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4(3):183–192
Yang MK, Ser SC, Lee CH (1989) Involvement of E. coli dcm methylase in Tn3 transposition. Proc Natl Sci Counc Repub China B 13(4):276–283
Yotani T, Yamada Y, Arai E, Tian Y, Gotoh M, Komiyama M, Fujimoto H, Sakamoto M, Kanai Y (2018) Novel method for DNA methylation analysis using high-performance liquid chromatography and its clinical application. Cancer Sci 109(5):1690–1700
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Chen, P., Bandoy, D.J.D., Weimer, B.C. (2020). Bacterial Epigenomics: Epigenetics in the Age of Population Genomics. In: Tettelin, H., Medini, D. (eds) The Pangenome. Springer, Cham. https://doi.org/10.1007/978-3-030-38281-0_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-38281-0_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-38280-3
Online ISBN: 978-3-030-38281-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)