Abstract
Adverse drug events (ADEs) are considered to be highly important and critical conditions, while accounting for around 3.7% of hospital admissions all over the world. Several studies have applied predictive models for ADE detection; nonetheless, only a restricted number and type of features has been used. In the paper, we propose a framework for identifying ADEs in medical records, by first applying the Boruta feature importance criterion, and then using the top-ranked features for building a predictive model as well as for clustering. We provide an experimental evaluation on the MIMIC-III database by considering 7 types of ADEs illustrating the benefit of the Boruta criterion for the task of ADE detection.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Adverse drug events (ADEs) refer to diagnoses corresponding to injuries that result from the use of a drug, including harm caused by the normal use of a drug, drug overdose, and use-related harms, such as from drug dose reductions and discontinuations of drugs administration [21]. ADEs possess high clinical relevance being that they account for approximately 3.7% of hospital admissions around the world [16]. Unfortunately, many ADEs are currently not being identified as such, due to limited knowledge about the effects of medical treatments, e.g., drugs being tested only in limited clinical trials under controlled conditions. An alternative approach is to resort to machine learning and the exploitation of the constantly growing amounts of information stored in electronic healthcare records (EHRs), so as to extract knowledge from past observations and learn how to identify new patient cases with a high risk of leading to an ADE.
With the adoption of EHRs, the amount of healthcare documentation is larger than ever, and there are several efforts underway to involve patients in their healthcare process through the use of patient generated data. Traditionally, data management and machine models have been developed by utilizing information from structured data fields [1, 14] as well as clinical text [12], little attention has been devoted to combining different data sources for the creation of richer overall models [33]. More importantly, these data sources are naturally characterized by high degree of sparsity and missing values. Consider for example a drug prescription variable (e.g., beta-blockers), which is typically administered to patients suffering from heart-related disorders. We should expect that this variable will be substantially empty for patients not suffering from any heart disease.
The problem of missing values in EHRs has been identified by several earlier studies [1, 3]. More recently, Bagattini et al. [1] propose three simple approaches for handling sparse features in EHRs for the task of ADE detection. Nonetheless, only one type of EHR features was used, corresponding to blood test measurements before the occurrence of an ADE; while diagnoses codes and drug prescriptions were excluded from the study. Moreover, the goal of that paper was to define simple temporal abstractions that take into account such temporal features with high degrees of sparsity. The objective of our study in this paper is to take a different research angle and approach the problem using feature importance to assess the statistical significance of multiple, heterogeneous EHR features in terms of their predictive performance. Moreover, we aim to define a more general approach to the problem of ADE detection in EHRs that can handle disparate feature types and, in the presence of sparse and noisy features, identify the subset of most significant class-distinctive features, that can then be used for both classification and clustering of ADEs in EHRs.
The main contributions of the paper include: (1) the formulation of a framework for identifying and assessing the importance of medical features in terms of their predictive performance, as well as their descriptive power for the problem of ADE detection; (2) the proposed framework employs the Boruta feature importance criterion as a first step, and then subsequently pipelines the selected features to building a predictive model for ADE prediction, as well as identifying clusters of patients under different ADE classes; (3) an extensive experimental evaluation on patient records obtained from the MIMIC databaseFootnote 1 including patients with 7 ADE types, and assessing (a) the predictive performance of four classification models using sets of features extracted by the Boruta criterion, as well as (b) the descriptive performance of clusters obtained using the highest scoring features in terms of the Boruta criterion under K-medoids.
2 Related Work
The wide usage of EHRs in medical research has recently increased the interest in the use of clinical data sources by medical practitioners as well as researchers from various fields [13, 32]. Numerous research directions arise for the problem of ADE identification, which is the key focus of this study [13]. Compared to traditional data sources, such as spontaneous reports [26], as well as other popular resources, such as social media data [28], EHRs contain data types and information that allow for incidence estimation and provide class labels for supervised machine learning. Research on mining both structured and unstructured EHR data for ADE detection is nascent, see e.g. [10, 11, 25, 29, 33].
The traditional approach for ADE identification is performed before the deployment of a drug. This is achieved by several rounds of clinical trials, which however are hampered by the fact that only a limited sample of patients is usually employed and monitored for a short or limited time period. Consequently, the phenomenon of ADE under-reporting arises as several serious ADEs are not detected during clinical trials but rather after the market deployment of a new drug. This typically results in having several drugs withdrawn. These limitations can be overcome by defining and employing rules for ADE detection [4, 8].
Machine learning is an alternative to ADE detection by the exploitation of rich data features in EHRs, such as for example blood tests [23]. More importantly, the development and application of machine learning models, both supervised and unsupervised, in a clinical setting can facilitate substantial improvements in terms of ADE detection while maintaining low hospitalization and treatment costs. We can identify four major lines of research on learning from EHRs [17]: (1) detection and analysis of comorbidities, (2) clustering patients with similar characteristics, (3) supervised learning, and (4) cohort querying and analysis. Examples of the above four categories are itemset mining, association rule extraction, and disproportionality analysis, prediction of critical healthcare and patient conditions, such as, for instance, smoking status quantification for a patient [31], patient safety and automated surveillance of ADEs [15], comorbidity and disease networks [4], processing of clinical text [11], identification of suitable individuals for clinical trials [24], as well as identification of temporal associations between medical events and first prescriptions of medicines for signaling the presence of an ADE [22].
3 The FISUL Framework
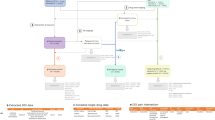
We present Feature Importance for Supervised and Unsupervised Learning (FISUL), a framework for predictive and descriptive modeling of ADEs from EHRs. FISUL has three phases: (1) feature importance, (2) predictive modeling, and (3) clustering. In Fig. 1 we provide an outline of the proposed framework. Next, we describe each phase in more detail.
3.1 Phase I: Boruta Feature Importance
We employ the Boruta method [19] as a feature importance criterion for reducing the number of data features. Boruta is a variable importance method that is defined for the random forest classifier, by mainly measuring the total decrease in impurity from performing a feature split over all nodes of a tree, averaged over all trees in the random forest. The Boruta method was selected for its ability to provide unbiased and stable selection of all relevant features [18].
The main idea is to create randomized copies of the existing features, merge the copies with the original data features, build a final classifier using all features, including the randomized ones, and iteratively identify the most important features for the classification task at hand. More concretely, let \(\mathcal {D}\) be the original dataset and let \(\mathcal {F}\) denote the original feature space. The key objective of Boruta is to define a mapping process, \(\mathcal {T}\), such that \(\mathcal {T}: \mathcal {F} \longrightarrow \hat{\mathcal {F}}\). Where \(\hat{\mathcal {F}}\) is a set of randomized features originating from \(\mathcal {F}\). More concretely, the following steps are performed:
-
Randomization: a replica, called shadow feature, \(\hat{f}_i\in \hat{\mathcal {F}}\) is created for each feature \(f_i\in \mathcal {F}\), by random permutation of its values; as a result, possible correlations that may exist between the original features and the class attribute are diminished.
-
Model building: a random forest \(\mathcal {R}\) is built using the union of the features \(\mathcal {F} \bigcup \hat{\mathcal {F}}\). This procedure is repeated n times, i.e., for n iterations.
-
Importance score: for each \(f_i\in \mathcal {F}\) and \(\hat{f}_i\in \hat{\mathcal {F}}\), we define an importance score, called Z-score, over all trees in \(\mathcal {R}\), where each feature appears. The mean and standard deviation of the accuracy loss are defined as \(\mu _{f_i}, \mu _{\hat{f}_i}\) and \(\sigma _{f_i}, \sigma _{\hat{f}_i}\), respectively, using the out-of-bag samples. Finally, the Z-score of each feature \(f_i\) and each shadow feature \(\hat{f}_i\) is defined as
$$ Z_{f_i} = \frac{\mu _{f_i}}{\sigma _{f_i}} \text { and } Z_{\hat{f}_i} = \frac{\mu _{\hat{f}_i}}{\sigma _{\hat{f}_i}}\ , $$respectively. Intuitively, the Z-score reflects the degree of fluctuation of the mean accuracy loss among the trees in \(\mathcal {R}\).
-
Statistical significance: for each original \(f_i\in \mathcal {F}\), we compute a statistical significance score using a two-tailed binomial test. More specifically, let \(Z^j_{max}\) be maximum Z-score of all shadow features in iteration j, i.e.,
$$\begin{aligned} Z^j_{max} = \max _{\hat{f}_i \in \hat{\mathcal {F}}} Z_{\hat{f}_i} \ . \end{aligned}$$(1)We use a vector, called hit vector \(\mathcal {H}\), to store for each \(f_i\in \mathcal {F}\) in how many iterations it achieved a Z-score higher than \(Z^j_{max}\), i.e.,
 (2)
(2)where
 is the indicator function, i.e.,
is the indicator function, i.e.,  (3)
(3)If feature \(f_i\) performs significantly better than expected compared to its shadow features in terms of Z-score, it is marked as “important”. Note that under the binomial distribution assumptions, the expected number of times \(\mathcal {H}_i\) that \(f_i\) may outperform its shadow replicas is simply \(E(f_i, n)= \frac{n}{2}\), with a standard deviation \(\sigma (f_i,n) = \sqrt{0.25 n}\), assuming that \(\mathcal {H}_i \sim B(n, 0.5)\). Conversely, \(f_i\) is considered “important”, if \(\mathcal {H}_i\) is significantly higher than \(E(f_i,n)\). Finally, the features that survive the significance test constitute the set of Boruta features \(\mathcal {F}^{\star }\).
3.2 Phase II: Predictive Modeling
The set of Boruta features \(\mathcal {F}^{\star }\) extracted from Phase I are next passed to Phase II for building a predictive model using the new feature space. The main objective is to learn a classification function \(\tau : o\rightarrow y\), that assigns a given data object o with a class label from a set \(\mathcal {C}\) of predefined class labels, such that \(y\in \mathcal {C}\). More specifically, we can couple \(\tau \) with a set of features \(\theta \) selected and employed during the training phase. In our case, the set of class labels corresponds to a selected set of ADEs. More information about the selected class labels can be found in Sect. 4. The training phase of a predictive model is more formally defined as \(\tau = \mathcal {L} (\theta , \mathcal {T})\), where \(\mathcal {L}\) is the learning function corresponding to a chosen predictive model and \(\mathcal {T}\) is the training set. Finally, the label of a newly seen data example o is obtained by applying \(\tau \), configured with the same chosen feature set \(\theta \), i.e., \(y = \tau (o;\theta )\). In our framework, we choose the top-k most important Boruta features, i.e., \(\theta = \mathcal {F}^{\star }_k\).
3.3 Phase III: Clustering
An alternative approach for exploiting \(\mathcal {F}^{\star }\) is clustering. The main objective is to define a partitioning \(\mathcal {G} = \{g_1, \ldots , g_K\}\) of K groups, such that inter-group similarity is maximized and intra-group similarity is minimized.
Since in our case the data objects contain features that are not necessarily numerical, we employ K-medoids using the Gower distance. This distance function computes the average dissimilarity across the data objects. Let \(o_i,o_j\) be two data objects in our dataset and \(|\theta |\) be the size of our feature space. The Gower distance is computed as follows:
where \(d^f_{i,j}\) is a function computing the dissimilarity of feature f between objects \(o_i\) and \(o_j\), depending on the feature type, after standardizing each feature. For example, in the case of numerical features, \(d^f_{i,j}\) is defined as follows:
where \(Z_f\) is the maximum distance range across all data objects and \(o^f_i,o^f_j\) denote the values of feature f for objects \(o_i, o_j\), respectively. In the case of categorical features, \(d^f_{i,j} =0\), if \( o^f_i=o^f_j\) and 1, otherwise.
The final clustering is obtained by running K-medoids under the Gower distance given by Eq. 4, and tuning K using the Silhouette coefficient [27] and selecting the one with the highest Silhouette value [27].
4 Experimental Evaluation
We outline the experimental setup by first our dataset, the benchmarked methods, the undersampling procedure we used to tackle the high class imbalance, and finally the presentation of our findings.
Dataset. We used the Medical Information Mark for Intensive Care III (MIMIC-III) database [30], a freely available medical database for intensive care (ICU) research, released in 2006 and comprising over 40,000 patients. Several studies have been conducted on this dataset using predictive models, such as prediction of hospital stay [9] or mortality rate [6]. However, little attention has been given to prediction of ADEs, yet they are common in ICU patients [2]. In MIMIC-III the ADEs are coded as ICD-9 diagnosis codes and for this study we explored the 7 of the most commonly occurring codes depicting ADEs; grouped as caused by one of four specific drugs: (1) antibiotics, (2) anticoagulants, (3) antineoplastic and immunosuppressive drugs, or (4) corticosteroids. We hence considered five datasets, one being the whole dataset including all ADEs, while each of the remaining four corresponded to each of the four specific drugs. All hospital admissions where one of these drugs were prescribed, were considered in the preprocessing, with a positive class label signifying at least one of the selected ADEs during hospital stay. A summary of the used datasets is given in Table 1.
Four different types of features were selected from MIMIC-III, either based on previous relevance to ADE prediction or as they had been identified clinically as risk factors or indications of ADEs in critical care. These features were: admission characteristics, undergone procedures, laboratory tests, and prescribed drugs. For the last three, one-hot encoding was applied based on clinically relevant groupings of their coding systems. The NDC drug codes extracted from MIMIC-III were converted to ATC codes [20], as the ATC grouping system has more clinical relevance. The full set of selected features is described in Table 2. The drug specific datasets excluded the drug feature group the ADE was caused by, for example the dataset with ADEs caused by corticosteroids excluded the corticosteroid drug group as a feature.
Setup. We benchmarked six predictive modeling techniques having demonstrated competitive predictive performance in earlier works on ADE detection [1, 33]: (1) Random Forests (RF100) with 100 trees, (2) simple Feed-Forward Neural Networks (NNet), (3) eXtreme gradient boosting (XGBoost), (4) SVM with a radial basis kernel (SVMRadial), (5) SVM with a polynomial kernel with degree 3 (SVMPolynomial), and (6) SVM with a linear kernel (SVMLinear). Due to the high class imbalance in all datasets, we performed under-sampling of the majority class for each dataset. All models used 3 feature sets: all features, the relevant features as selected by Boruta, and Boruta’s top 10 (after under-sampling). The performance metrics were AUC and AUPRC, under 10-fold cross-validation. For clustering we used the original imbalanced datasets. We applied K-medoids for different values of K, using the Gower distance on the top-10 Boruta selected features.
Results. Next, we present our experimental findings for each of the three phases of the FISUL framework.
-
Boruta feature importance. When applied to the whole dataset, the Boruta criterion rejected 56 features, mainly those that were extremely sparse (<1% occurrence). Many of the top ranking significant features are laboratory tests, while the vast majority of the remaining significant features are drug prescriptions with very few admission characteristics and procedures, which is comparable to the antibiotics dataset (Fig. 2). The anticoagulants dataset paints a similar picture, with the only difference that age and the platelets lab test are the top 2 features (Fig. 4). For the immunosuppressives dataset, the top-3 features were again laboratory tests, while the majority of the remaining significant features were drugs, with very few features corresponding to procedures. On the other hand, we observe that for corticosteroids (Fig. 3 (left)) the vast majority of significant features are drugs, with laboratory tests being less significant. We also note that for the latter dataset, no other feature types where deemed significant.
-
Predictive modeling. Our experimental findings in terms of predictive performance are shown in Tables 3, 4, and 5. We observe that both RF100 and XGBoost consistently outperform the other classifiers for the whole dataset classification in all feature sets. For the four individual datasets, RF100 is still a winner in most of the cases, alongside with XGBoost, especially on the complete unfiltered feature set, where the other benchmarked classifiers are not competitive. However, note that when the Boruta feature selection is applied first and either only all relevant or top-10 features are included, the performances of the other benchmarked classifiers are not substantially lower than the two winners. Clustering. For all 5 datasets, the Silhouette coefficient suggested two clusters. For all cases, the two clusters mostly contained non-ADE examples, while cluster purity was above 96% for the whole dataset as well as for corticosteroids, 98% for antibiotics and anticoagulants, and 91% for immunosuppressives. Due to the inherent extreme class imbalance of the dataset the top-10 Boruta features did not manage to capture any strong cluster structure that can distinguish ADE from non-ADE cases.
Histograms of the Boruta importance for all feature types for the corticosteroids dataset (left) and the immunosuppressive dataset (right). We observe that the features with the highest importance for immunosuppressives are the drugs, while for corticosteroids some laboratory tests precede in the ranking.
Discussion. Our overall framework and our benchmark on six predictive models, illustrates the medical importance of the selected features. The most important features of all the datasets mainly included values clinically known to be indicative of ADEs, such as laboratory tests and certain drugs groups. The procedures seemed to be the least influential features for ADE detection. There were differences between the 4 drug-specific datasets, which could be clinically explained. For example, the top 2 features in the anticoagulants dataset were age and platelet lab tests, which are also considered the two major risk factors in anticoagulants caused ADEs [7]. Moreover, the corticosteroid dataset showed mainly drugs important features. Corticosteroids are a known factor in many drug-drug interactions [5], which are often at the base of ADEs, which could explain the importance of other drug groups for corticosteroid-induced ADEs.
With regards to the classifiers, it is evident that the random forest and extreme gradient boosting outperformed the other classifiers. However, when the full framework including the feature selection is applied, the other classifiers become competitive, while the two winners to not decrease in predictive performance. Also, the addition of the feature selection speeds up the model building phase substantially, without decreasing the performance. Moreover, due to the high values of purity obtained in the clustering, our results still suggest that the Boruta criterion can be seen as a promising feature importance measure for identifying strong cluster substructures.
5 Conclusions
We presented a framework for studying ADEs in EHRs using heterogeneous feature types. We illustrated the importance of the Boruta feature importance criterion for the tasks of classification and clustering ADEs. Our findings suggest that integrating different feature types along with a strong feature importance criterion (such as Boruta) can provide substantially better predictive performance compared to only using drugs or clinical tests. Directions for future work include the integration of features from non-structured data types (e.g., clinical text and notes) and the exploration of alternative feature importance measures.
Notes
References
Bagattini, F., Karlsson, I., Rebane, J., Papapetrou, P.: A classification framework for exploiting sparse multi-variate temporal features with application to adverse drug event detection in medical records. BMC Med. Inform. Decis. Making 19(1), 7 (2019)
Bates, D.W., et al.: Patient risk factors for adverse drug events in hospitalized patients. Arch. Intern. Med. 159(21), 2553–2560 (1999)
Beaulieu-Jones, B.K., Lavage, D.R., Snyder, J.W., Moore, J.H., Pendergrass, S.A., Bauer, C.R.: Characterizing and managing missing structured data in electronic health records: data analysis. JMIR Med. Inform. 6(1), e11 (2018)
Cao, H., Markatou, M., Melton, G.B., Chiang, M.F., Hripcsak, G.: Handling temporality of clinical events for drug safety surveillance. In: AMIA Proceedings, vol. 2005, pp. 106–110. American Medical Informatics Association (2005)
Daveluy, A., Raignoux, C., Miremont-Salamé, G., Girodet, P., Moore, N., Haramburu, F., Molimard, M.: Drug interactions between inhaled corticosteroids and enzymatic inhibitors. Eur. J. Clin. Pharmacol. 65(7), 743–745 (2009)
Desautels, T., et al.: Using transfer learning for improved mortality prediction in a data-scarce hospital setting. Biomed. Inform. Insights 9, July 2017
Fitzmaurice, D., Blann, A., Lip, G.: Bleeding risks of antithrombotic therapy. Br. Med. J. 325(7368), 828–831 (2002)
Freeman, R., Moore, L., García Alvarez, L., Charlett, A., Holmes, A.: Advances in electronic surveillance for healthcare-associated infections in the 21st century: a systematic review. J. Hosp. Infect. 84(2), 106–119 (2013)
Gentimis, T., Alnaser, A.J., Durante, A., Cook, K., Steele, R.: Predicting hospital length of stay using neural networks on mimic iii data. In: 2017 IEEE 15th Intl Conf on Dependable, Autonomic and Secure Computing, 15th Intl Conf on Pervasive Intelligence and Computing, 3rd Intl Conf on Big Data Intelligence and Computing and Cyber Science and Technology Congress (DASC/PiCom/DataCom/CyberSciTech), pp. 1194–1201, November 2017
Harpaz, R., Haerian, K., Chase, H.S., Friedman, C.: Mining electronic health records for adverse drug effects using regression based methods. In: The 1st ACM International Health Informatics Symposium, pp. 100–107. ACM (2010)
Henriksson, A., Kvist, M., Dalianis, H., Duneld, M.: Identifying adverse drug event information in clinical notes with distributional semantic representations of context. J. Biomed. Inform. 57, 333–349 (2015)
Henriksson, A., Zhao, J., Boström, H., Dalianis, H.: Modeling heterogeneous clinical sequence data in semantic space for adverse drug event detection. In: IEEE International Conference on Data Science and Advanced Analytics, pp. 1–8 (2015)
Hersh, W.R.: Adding value to the electronic health record through secondary use of data for quality assurance, research, and surveillance. Clin. Pharmacol. Ther. 81, 126–128 (2007)
Hielscher, T., Spiliopoulou, M., Völzke, H., Kühn, J.: Mining longitudinal epidemiological data to understand a reversible disorder. In: International Symposium on Intelligent Data Analysis, pp. 120–130 (2014)
Honigman, B., et al.: Using computerized data to identify adverse drug events in outpatients. J. Am. Med. Inform. Assoc. 8(3), 254–266 (2001)
Howard, R., Avery, A., Slavenburg, S., Royal, S., Pipe, G., Lucassen, P., Pirmohamed, M.: Which drugs cause preventable admissions to hospital? a systematic review. Br. J. Clin. Pharmacol. 63(2), 136–147 (2007)
Jensen, P.B., Jensen, L.J., Brunak, S.: Mining electronic health records: towards better research applications and clinical care. Nature Rev. Genet. 13(6), 395–405 (2012)
Kursa, M., Rudnicki, W.: Feature selection with the boruta package. J. Stat. Softw. 36(11), 1–13 (2010)
Kursa, M.B., Jankowski, A., Rudnicki, W.R.: Boruta - a system for feature selection. Fundam. Inf. 101(4), 271–285 (2010)
Kury, F., Bodenreider, O.: Desiderata for drug classification systems for their use in analyzing large drug prescription datasets. In: Proceedings of the 3rd Workshop on Data Mining for Medical Informatics (2016)
Nebeker, J.R., Barach, P., Samore, M.H.: Clarifying adverse drug events: a clinician’s guide to terminology, documentation, and reporting. Ann. Internal Med. 140(10), 795–801 (2004)
Norén, G.N., Bergvall, T., Ryan, P.B., Juhlin, K., Schuemie, M.J., Madigan, D.: Empirical performance of the calibrated self-controlled cohort analysis within temporal pattern discovery: lessons for developing a risk identification and analysis system. Drug Saf. 36(1), 107–121 (2013)
Ouchi, K., Lindvall, C., Chai, P.R., Boyer, E.W.: Machine learning to predict, detect, and intervene older adults vulnerable for adverse drug events in the emergency department. J. Med. Toxicol. 14(3), 248–252 (2018)
Pakhomov, S.V., Buntrock, J., Chute, C.G.: Prospective recruitment of patients with congestive heart failure using an ad-hoc binary classifier. J. Biomed. Inform. 38(2), 145–153 (2005)
Park, M.Y., et al.: A novel algorithm for detection of adverse drug reaction signals using a hospital electronic medical record database. Pharmacoepidemiol. Drug Saf. 20(6), 598–607 (2011)
van Puijenbroek, E.P., Bate, A., Leufkens, H.G., Lindquist, M., Orre, R., Egberts, A.C.: A comparison of measures of disproportionality for signal detection in spontaneous reporting systems for adverse drug reactions. Pharmacoepidemiol. Drug Saf. 11(1), 3–10 (2002)
Rousseeuw, P.J.: Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987)
Sarker, A., et al.: Utilizing social media data for pharmacovigilance: a review. J. Biomed. Inform. 54, 202–212 (2015)
Schuemie, M.J., et al.: Using electronic health care records for drug safety signal detection: a comparative evaluation of statistical methods. Med. Care 50(10), 890–897 (2012)
Scott, D.J., et al.: Accessing the public mimic-ii intensive care relational database for clinical research. BMC Med. Inform. Decis. 13(1), 9 (2013)
Uzuner, Ö., Goldstein, I., Luo, Y., Kohane, I.: Identifying patient smoking status from medical discharge records. J. Am. Med. Inform. Assoc. 15(1), 14–24 (2008)
Weiskopf, N.G., Hripcsak, G., Swaminathan, S., Weng, C.: Defining and measuring completeness of electronic health records for secondary use. J. Biomed. Inform. 46(5), 830–836 (2013)
Zhao, J., Henriksson, A., Asker, L., Boström, H.: Predictive modeling of structured electronic health records for adverse drug event detection. BMC Med. Inform. Decis. Making 15(Suppl 4), S1 (2015)
Acknowledgments
This work was partly supported by the VR-2016-03372 Swedish Research Council Starting Grant.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 IFIP International Federation for Information Processing
About this paper
Cite this paper
Allaart, C.G., Mondrejevski, L., Papapetrou, P. (2019). FISUL: A Framework for Detecting Adverse Drug Events from Heterogeneous Medical Sources Using Feature Importance. In: MacIntyre, J., Maglogiannis, I., Iliadis, L., Pimenidis, E. (eds) Artificial Intelligence Applications and Innovations. AIAI 2019. IFIP Advances in Information and Communication Technology, vol 559. Springer, Cham. https://doi.org/10.1007/978-3-030-19823-7_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-19823-7_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-19822-0
Online ISBN: 978-3-030-19823-7
eBook Packages: Computer ScienceComputer Science (R0)






 is the indicator function, i.e.,
is the indicator function, i.e., 



