Abstract
Since the introduction of BOLD (Blood Oxygen Level Dependent) imaging, the hemodynamic response model has remained the standard analysis approach to relate activated brain areas to extrinsic task conditions. Ongoing brain activity unrelated to the task is neglected and considered noise. By contrast, model-free blind source separation techniques such as Independent Component Analysis (ICA) have been used in intrinsic task-free experiments to reveal functional systems usually referred to as “resting-state” networks. However, matrix factorization techniques applied to BOLD imaging do not model the translation of neuronal activity into BOLD fluctuations and depend on arbitrarily chosen regularization measures such as statistical independence or sparsity. We present a novel neurobiologically-driven matrix factorization approach. Our matrix factorization model incorporates the hemodynamic response function that enables the estimation of underlying neural activity in individual brain networks that present during task- and task-free BOLD-fMRI experiments. We validate our model on the recently published Midnight Scanning Club dataset including five hours of task-free and six hours of various task experiments per subject. The resulting temporal and spatial activation patterns obtained from our matrix factorization technique resemble individual task profiles and known functional brain networks, which are either correlated with the task or spontaneously activating unrelated to the task.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
The most common experimental approach in BOLD-fMRI is to design task experiments that stimulate individual brain areas causing increased local neuronal activity and changes in local blood flow. A binary function of neural activity comprises onset and duration of the task, and is convolved with a hemodynamic response function (HRF), which relates neural activity to hemodynamic changes. On the basis of this established link between neuronal activity and BOLD signal, several task experiments have been developed to map out underlying functional systems in humans. By contrast, ongoing spontaneous neural activity from task-free fMRI experiments have been found to organize into so called resting-state networks of intrinsic functional connectivity. In contrast to task-fMRI, model-free approaches like Independent Component Analysis (ICA) or seed-based analysis have been used to study this intrinsic functional connectivity, but these do not model the relation between underlying neuronal activity and spontaneous BOLD signal fluctuations. Recent studies [1] that investigated the link between resting-state and task-fMRI have provided evidence that there is a common functional brain architecture that exhibits increased or decreased neural activity dependent on the intrinsic or extrinsic task demands. We propose a novel generative model embedded within a neural network optimization framework to obtain characteristic brain networks and their corresponding neural activation profiles during a given task. The framework only requires minimal pre-processing and is regularized by the known HRF.
2 Materials and Methods
We propose a Convolutional Hemodynamic Autoencoder (CHA) that results in components which we loosely associate with functional networks. Each functional network consists of three parts: a spatial map; a neuronal activity time course; and a corresponding BOLD time course. We test our proposed technique on both simulated and real imaging data with minimal preprocessing.
2.1 Functional Imaging Data
We use simulated data fromFootnote 1 and compare our results to the method of Total Activation (TA) [2]. We further evaluate our method on the Midnight Scanning club (MSC) dataFootnote 2 that comprises 10 healthy adult subjects with six hours of task-free and five hours of task-based BOLD-fMRI experiments including motor tasks, incidental memory and a mixed design task. More details can be found in the respective references [2, 3].
2.2 Minimal Preprocessing
For the fMRI sequences of each subject, volumes were realigned to the first volume to correct for head motion. The first volume was linearly registered to its corresponding bias-corrected T1-weighted anatomical scan. The intra-subject affine registration and non-linear registration to the Talairach template were combined to map all fMRI volumes with one re-sampling into the Talairach space sampled in a 4 mm isotropic resolution. Time courses of voxels within brain tissue were extracted. Time courses were high-pass filtered (0.01 Hz cut-off) to remove signal drifts from scanner instabilities. Time courses are centered and variance-normalized. The functional 4D volume set per subject is reshaped into a matrix \(X \in \mathbb {R}^{t\times v}\) with t time points and v voxels.
2.3 Generative Model
Our CHA assumes that BOLD-fMRI data can be modeled as a compressed representation of c components. Each component i consists of a spatial map \(h_i \in \mathbb {R}^{1\times v}_+\) of v voxels, a neural activity time course \(N_i \in \mathbb {R}^{t\times 1}_+\) of t time points. The corresponding BOLD time-course \(W_i=N_i\circledast \mathcal {H}\) is obtained by convolving \(N_i\) with an impulse response function \(\mathcal {H}\) (the standard HRF shape model obtained from two Gamma functions). This generative model in matrix notation is given by:
with observed BOLD time course matrix \(X \in \mathbb {R}^{t\times v}\) decomposed into BOLD time course matrix \(W \in \mathbb {R}^{t\times c}\) and spatial component matrix \(h \in \mathbb {R}^{c\times v}_+\). Each BOLD time course in W has a corresponding time course in the neural activity time course matrix N. Bias \(b_2 \in \mathbb {R}\) creates a negative baseline to compensate for negative time course values after preprocessing described in Sect. 2.2.
2.4 Convolutional Hemodynamic Autoencoder
The generative model is embedded into a neural network autoencoder framework. The encoder maps X into the hidden layer \(h=f({W^T}X+{b_1})\) using a rectified linear unit (ReLU) activation function \(f:\mathbb {R}\rightarrow \mathbb {R}_+\), activation bias \({b_1} \in \mathbb {R}^{1\times t}\) and the transpose of BOLD time course matrix W introduced in Sect. 2.3. The parameters of the autoencoder are neural activity time courses N and biases \(b_1\) and \(b_2\) resulting in the following cost function:
minimizing the l2 norm between the original data X and its reconstruction \(\hat{X}\). Neural activity time courses N are initialized with the absolute values of a random Xavier initialization [4]. Biases \(b_1\) and \(b_2\) are initialized with zeros. The back-propagation algorithm (chain rule) is applied to derive the corresponding gradient for the cost function. The gradient is optimized with the limited memory Broyden-Fletcher-Goldfarb-Shanno (L-BFGS) optimization scheme.
2.5 Hyper-parameter Tuning
The only hyper-parameter to tune is the number of components c. We use the following heuristic to find a good solution through model averaging. We run n = 5 random initializations with \(c=1000\) for each of the 10 subjects (\(\#subj\)) per taskFootnote 3. We apply a non-negative matrix factorization (NMF) with non-negative double singular value initialization (NNDSVD) [5] to cluster all combined spatial maps \(H \in \mathbb {R}^{(c\times n\times \#subj)\times v}_+\). The NNDSVD speeds up convergence and guarantees deterministic behavior for determining the right number of components \(c_{opt}\) in the following NMF cross-validation with Gabriel holdout as described in [6]. The complete processing pipeline is summarized in Fig. 1. Subsequently, subject specific neural activity and BOLD time courses as well as spatial maps are obtained by weighted average informed by the association matrix W.
For each task, we concatenate all session data and compute n CHA runs for each subject. We obtain \(c\times n\times \#subj\) components. Each component consists of a neural activity time course, BOLD time course and spatial map. We cluster all spatial maps with a NMF and cross-validate to obtain an optimal decomposition with \(c_{opt}\) per task. We use the individual weights of the association matrix W to compute a weighted average of neural activity time course, BOLD time course and spatial maps per component.
3 Results
3.1 Simulation Data
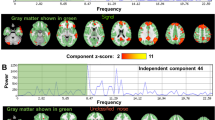
Figure 2 shows the results from simulated data; the simulation data contains four components (top left) and their corresponding neural activity time courses (bottom row). Cross-validation (bottom left) results in a very similar error for four and more components. We obtained neural activity time courses and spatial maps using \(c_{opt}=4\). Our technique is able to find a close estimate of the correct dimensionality of the decomposition and recovers the respective time course and spatial map of each component successfully.
3.2 Imaging Data
We obtain an optimal group decomposition into \(c_{opt}\) components for each task that provides weighted averages of the neural activity time course and spatial map per component in each individual subject as outlined in Fig. 1. For example, the optimal value of \(c_{opt}\) for the motor task is 50 components determined by cross-validation with 9-fold Gabriel holdout as depicted in Fig. 3. Six of the 50 components with their corresponding spatial map, neural activation and BOLD time course for subject one are depicted for the motor task in Fig. 4. All first sessions for all three tasks for each subject are included in the Supplementary Material. We found five activated brain networks occurring in all tasks. The Default Mode Network (DMN), Visual Network, Precuneus Network, Salient Network, Right and Left Central Executive Network (RCEN and LCEN). We examined the correlation of the neural activation time course in these five networks with the visual cues in each task for all sessions summarized in box-plots depicted in Fig. 3. The neural activation in the Visual Network follows the visual cues with a small delay as seen in Fig. 4. The Salient Network also exhibits minor correlation to the visual stimuli, while most other networks, such as the DMN, express unrelated neural activity to the task experiment.
We conducted a 9-fold cross-validation with Gabriel holdout to determine an optimal decomposition for each individual task that fits all subject sessions. The three figures on the right depict the correlation of the neural activation time course of common brain networks to the visual cues in motor, memory and glass-lexical task among all sessions and subjects.
The first four activated brain regions belong to the Sensory-Motor (SMN) network and relate to left and right foot, tongue, left and right hand stimuli, respectively. The fifth and sixth brain network are Visual Network (VN) and Default Mode Network (DMN), respectively. The blue lines represent parts of the block design of the motor task. The red and green line respectively represent neural activity and BOLD signal change in each individual network. We observe that the first five networks follow the respective block stimuli or visual cues given during the task. In contrast, the DMN network exhibits a neural activation profile unrelated to the task but remains strongly detectable.
4 Discussion
We have presented a new technique that simultaneously decomposes the observed BOLD signal into maps of spatial position, neuronal activity time courses, and hemodynamic responses.
This has several advantages over existing methods. In task-based fMRI the general linear model relates the experiment time course to observed BOLD fluctuations, but does not allow for ongoing spontaneous BOLD fluctuations unrelated to the task. Furthermore, it models underlying neural activity as switching from off to on instantly. A comprehensive GLM analysis of the several contrasts for the MSC dataset is availableFootnote 4. Our generative model estimates the underlying neuronal activity time course and does not know the underlying experimental design and it is therefore truly unsupervised. When compared to resting-state fMRI, matrix decomposition techniques are often used to extract brain networks [7, 8]. The disadvantage of these techniques is that they require certain statistical assumptions to hold for the underlying sources of interest. Although effective at removing scanner- or physiological motion-related noise sources, they approximate statistical independence with non-linear functions. The chosen non-linearity has a strong influence on the obtained value distribution of voxel intensities and thus its spatial characteristics in the case of BOLD-fMRI. Additionally, the seminal work by [9] has shown that the commonly used algorithms in ICA, FastICA and InfoMax, tend to produce sparse rather than independent sources in simulated BOLD-fMRI data. This has shifted the focus in the recent years to sparse decomposition techniques [8, 9] with weaker model assumptions. However, the these techniques require regularization and therefore hyper-parameter tuning with some sort of cross-validation, which is computationally expensive or intractable depending on the number of parameters to tune. For example, the most similar approach [2] to our proposed CHA generative model requires to tune a spatial and a temporal regularization hyper-parameter. Given that a 5 mm spatial smoothing kernel was applied, the value of the spatial regularization hyper-parameter is weakly motivated and could supposedly not be cross-validated due to the long individual subject processing time (5 h). The spatial smoothing kernel size hyper-parameter varies in studies between 3 to 12 mm. The kernel size thus determines partial-volume effects at both the intra- and inter-individual level. Unique subject network representations are therefore lost or compromised.
Combined, these limitations of common pre- and post-processing techniques potentially make subsequent analysis incomparable. Our proposed generative model delivers smooth spatial network maps with minimal pre-processing and without spatial regularization. Our technique exploits what is known about the neurovascular coupling between neuronal activity and blood perfusion. It does not require artificial spatial or temporal regularization but leverages biological prior information. The decomposition of a BOLD-fMRI scan into spatial network maps, functional time-courses and hemodynamic responses opens the door to sophisticated analyses at both group and single-subject level to examine neural activity in both, spontaneously activating networks and networks engaged in an extrinsic task. The obtained neural activity profile per network provides a means to quantify the activity during a 5 to 10-min scan, opening new analysis routes to clinical diagnosis and drug testing in neurological disorders.
Our technique relies on finding an optimal number of components similar to all matrix factorization techniques. This is challenging because the brain will form networks dynamically and dependent on the task. We find a good approximation of individual interacting networks during task and rest by applying sophisticated cross-validation and model averaging strategies.
Our proposed Convolutional Hemodynamic Autoencoder will therefore provide new insights about the underlying cause of BOLD signal change in task and task-free BOLD-fMRI in current and future large fMRI cohort studies.
Notes
- 1.
- 2.
- 3.
Limited by the available 24 GB GPU memory.
- 4.
References
Tavor, I., Jones, O.P., Mars, R., Smith, S., Behrens, T., Jbabdi, S.: Task-free MRI predicts individual differences in brain activity during task performance. Science 352(6282), 216–220 (2016)
Karahanoğlu, F.I., Caballero-Gaudes, C., Lazeyras, F., Van De Ville, D.: Total activation: fMRI deconvolution through spatio-temporal regularization. Neuroimage 73, 121–134 (2013)
Gordon, E.M., et al.: Precision functional mapping of individual human brains. Neuron 95(4), 791–807 (2017)
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, pp. 249–256 (2010)
Boutsidis, C., Gallopoulos, E.: Svd based initialization: a head start for nonnegative matrix factorization. Pattern Recognit. 41(4), 1350–1362 (2008)
Kanagal, B., Sindhwani, V.: Rank selection in low-rank matrix approximations: a study of cross-validation for NMFs. In: Proceedings of the Conference on Advance Neural Information Processing, vol. 1, pp. 10–15 (2010)
Beckmann, C.F., Mackay, C.E., Filippini, N., Smith, S.M.: Group comparison of resting-state fMRI data using multi-subject ICA and dual regression. Neuroimage 47(Suppl. 1), S148 (2009)
Abraham, A., Dohmatob, E., Thirion, B., Samaras, D., Varoquaux, G.: Extracting brain regions from rest fMRI with total-variation constrained dictionary learning. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013. LNCS, vol. 8150, pp. 607–615. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-40763-5_75
Daubechies, I., et al.: Independent component analysis for brain fmri does not select for independence. Proc. Natl. Acad. Sci. 106(26), 10415–10422 (2009)
Acknowledgements
We would like to acknowledge the Wellcome Trust (210182/Z/18/Z, 101957/Z/13/Z), EPSRC (NS/A000027/1), MRC (MR/J01107X/1), and the Wolfson Foundation (PR/ylr/18575).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Hütel, M., Melbourne, A., Ourselin, S. (2018). Neural Activation Estimation in Brain Networks During Task and Rest Using BOLD-fMRI. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11072. Springer, Cham. https://doi.org/10.1007/978-3-030-00931-1_25
Download citation
DOI: https://doi.org/10.1007/978-3-030-00931-1_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00930-4
Online ISBN: 978-3-030-00931-1
eBook Packages: Computer ScienceComputer Science (R0)