Abstract
In this paper, we propose an order-sensitive deep hashing for scalable medical image retrieval in the scenario of coexistence of multiple medical conditions. The pairwise similarity preservation in existing hashing methods is not suitable for this multimorbidity medical image retrieval problem. To capture the multilevel semantic similarity, we formulate it as a multi-label hashing learning problem. We design a deep hash model for powerful feature extraction and preserve the ranking list with a triplet based ranking loss for better assessment assistance. We further introduce the cross-entropy based multi-label classification loss to exploit multi-label information. We solve the optimization problem by continuation to reduce the quantization loss. We conduct extensive experiments on a large database constructed on the NIH Chest X-ray database to validate the efficacy of the proposed algorithm. Experimental results demonstrate that our order sensitive deep hashing leads to superior performance compared with several state-of-the-art hashing methods.
Z. Chen and R. Cai—Co-first authors.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
The pictures of internal body structures produced by CT and MRI scans are important for the diagnosis and assessment of disease. The interpretation of the imaging results is objective and with high inter-observer variability due to the requirement of expertise accumulation and practical experience. To circumvent the discrepancy between expert interpretations, prior cases with similar manifestations could be presented to form a reference based assessment by content based image retrieval. For better assistance in assessment, such retrieval system should be with plenty cases of various disease manifestations, which in turn requires the similar retrieval algorithm to be both scalable and accurate. Learning based hashing methods arise to be a promising solution for such retrieval system by encoding images as compact binary codes with similarity preservation in the Hamming space [1].
Learning based hashing methods leverage the statistical properties of data samples to learn the mapping functions to generate compact binary codes. They can be broadly categorized into shallow learning based hashing methods and deep learning based hashing methods. The former takes handcrafted features like SIFT and GIST as input and learns hashing functions to transform them into compact binary codes. Representative works in this class includes Spectral Hashing (SH) [2] that solves eigenvectors of the graph Laplacian with bit balance and bit independent constraints, Iterative Quantization (ITQ) [3] that further improves the results by reducing the quantization loss through feature rotation, Semi-supervised Hashing (SSH) [4] that exploits both the unlabelled and labelled data. They learn the hashing functions in a two stage manner to optimize transformations with feature fixed, which may lead to suboptimal performance. In contrast, deep learning based hashing methods are able to tailor features for hashing through end-to-end learning on the images directly and further enhance the performance with powerful convolutional neural network. The seminal work includes Deep Hashing (DH) [5] that utilizes multi-layer neural network to capture the nonlinear neighborhood relationship between samples, Deep Supervised Hashing (DSH) [6] that introduces a regularizer to encourage outputs of neural networks to be close to binary values, HashNet [7] that continuously approximates the sign activation with smooth activations. This motivates us to leverage the deep learning framework for hashing function learning.
For similarity preservation, the objective function of hash learning, both shallow and deep learning based hashing methods, is designed to align the distances or similarities computed from the input space and the Hamming space. The alignment is usually measured over a pair of samples with discrepancy minimization [8], such as the similarity-distance production minimization in spectral hashing. The pairwise distance in the Hamming space is desired to be smaller if the pairwise similarity in the input space is larger. Such similarity preservation is also used to develop the application specific hashing methods in the community of medical image computing, such as Deep Multiple Instance Hashing for tumor assessment [9], binary code tagging and Deep Residual Hashing for chest X-ray images [10, 11], etc. Note that such similarity preservation is suitable for samples with single class label. However, in the scenario of medical image, multiple symptoms or diseases may be observed from one medical image. Multilevel semantic structural similarity exists between samples, which the above pairwise alignment cannot capture. To this end, it is important to design objective function with multilevel similarity preservation in parallel to these existing methods.
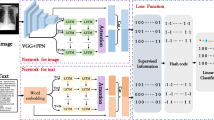
In this work, we propose an order sensitive deep hashing (termed as OSDH) method for scalable medical image retrieval with multimorbidity awareness, as shown in Fig. 1. We formulate this multimorbidity aware retrieval as a multi-label hash learning problem and leverage the convolutional neural network for feature extraction. We propose to solve it by optimizing the objective of triplet based ranking similarity preservation over binary codes. We further narrow the semantic gap between learned binary codes and the associated concepts with classification supervision. We apply the proposed OSDH algorithm to clinical chest X-ray database to validate the efficacy and demonstrate superior performance over several state-of-the-art hashing methods.
2 Methodology
Mathematically, given a set of training samples \({\varvec{X}}=\{{{\varvec{x}}}_1,\dots ,{{\varvec{x}}}_N\}\) and corresponding class labels \({\varvec{L}}=\{1,\dots ,C\}\), where each sample \({{\varvec{x}}_i}\) is associated with a subset of labels \({\varvec{Y}}_i\subseteq {\varvec{L}}\), our goal is to learn the hash functions to generate binary codes \({\varvec{B}}=\{{{\varvec{b}}}_1,\dots ,{{\varvec{b}}}_N\}\in \{-1,1\}^k\) such that the multilevel semantic structural similarity of samples is preserved by the binary codes. For scalable retrieval, the length of binary code k is much smaller than the dimension of input sample.
2.1 Deep Hash Model
As shown in Fig. 1, we develop a deep hash model to jointly learn visual feature extraction and the subsequent mapping to compact binary codes. The learning procedure is applied on raw pixels of input images by using convolutional neural network for feature extraction. Such hierarchical non-linear function exhibits powerful learning capacity and encourages the learned feature to capture the multilevel semantic information. The convolutional neural network could be an off-the-shelf architecture, such as AlexNet [12] or an application specific network. On top of the network, the output of the last fully connected layer \({\varvec{h}}_i\) is fed into the succeeding hash layer for dimensional reduction and binarization. We leverage a fully connected layer to map \({\varvec{h}}_i\) to a k-dimension feature vector \(\hat{\varvec{h}}_i^k\). \(\hat{\varvec{h}}_i^k\) is then quantized to \([-1,1]\) to produce the binary code \({\varvec{b}}_i\). To reduce the quantization loss, \(\hat{\varvec{h}}_i^k\) is usually passed through an activation layer to scale the magnitude within \([-1,1]\) before applying the binarization. While most existing works use the hyperbolic tangent function \(\text {tanh}(\hat{\varvec{h}}_i^k)\) in the activation layer, we design a parameterized hyperbolic tangent function \(\text {tanh}(\alpha \hat{\varvec{h}}_i^k)\) to approximate the \(\text {sgn}(\cdot )\) function, as will be detailed in Sect. 2.3. By denoting the mapping from raw pixels of image \({\varvec{x}}_i\) to the output of activation \(\text {tanh}(\alpha \hat{\varvec{h}}_i^k)\) as \({\varvec{g}}(\cdot )\) and its parameters as \(\varTheta \), we can formulate the derivation of binary code as
2.2 Order Sensitive Supervision
To facilitate efficient multimorbidity aware retrieval, the learned binary codes are expected to preserve the multilevel semantic similarity between samples. In the context of multiple labels, the similarity between samples can be measured by the ranking order of neighbors. For each query sample \({\varvec{x}}_q\), its semantic similarity level r with respect to a sample \({\varvec{x}}_i\) in the database can be computed by the number of common labels shared by both \(|{\varvec{Y}}_q\cap {\varvec{Y}}_i|\). By assigning a similarity level for each sample in the database, a ground truth ranking list for \({\varvec{x}}_q\) can be formed by sorting samples in the decreasing order of similarity level. For each query \({\varvec{x}}_q\) and its corresponding ranking list \(\{{\varvec{x}}_i\}_{i=1}^{M}\), we can define a triplet based ranking loss over binary codes,
\(D({\varvec{b}}_1,{\varvec{b}}_2)\) measures the Hamming distance between the binary codes \({\varvec{b}}_1\) and \({\varvec{b}}_2\). \(\rho \) is introduced to control the minimum margin between the Hamming distances of the two pairs. \(r_i\) and \(r_j\) are the ground truth similarity levels of samples \({\varvec{x}}_i\) and \({\varvec{x}}_j\) with respect to query \({\varvec{x}}_q\). Z is a constant related to the length of ranking list, which will be explained in Sect. 3. The coefficient \({\frac{2^{r_i}-2^{r_j}}{Z}}\) assigns larger weight for pair \(({\varvec{x}}_i,{\varvec{x}}_j)\) when \({\varvec{x}}_i\) is more relevant to \({\varvec{x}}_q\) than \({\varvec{x}}_j\). By summing over all the samples \({\varvec{x}}_i\) in the ranking list and its pair \(({\varvec{x}}_i,{\varvec{x}}_j)\), the minimization of (2) is able to encourage the preservation of the ranking list in the Hamming space for query \({\varvec{x}}_q\). To preserve the semantic multilevel similarity structure, we can choose to optimize the summation of (2) over all training samples, \(\sum _{{\varvec{x}}_q\in {\varvec{X}}}\mathcal {L}_R({\varvec{x}}_q)\).
While the loss in (2) is related to the relative similarity level, the label information is not fully exploited to learn hash functions. Previous works on single label data further take advantage of the label information by directly applying it to train the network [13, 14]. The training procedure is performed either in the framework of two-stream multi-task learning including classification and hash or by classification over the binary codes directly. The basic assumption of such algorithm is that the binary codes should be ideal for classification. In order to further exploit the multi-label information, we choose to expect the activation output \({\varvec{g}}({\varvec{x}}_i,\varTheta )\) optimal for classification and jointly learn both the network and the classifier. Specifically, we design the loss of multi-label classification in the form of cross entropy,
The ground truth label \({\varvec{y}}_{ic}\in \{0,1\}\) indicates whether sample \({\varvec{x}}_i\) is with the c-th label. For sample \({\varvec{x}}_i\), the probability belonging to the c-th class inferred by a linear classifier. By accumulating the cross-entropy loss of each class, (3) presents the multi-label classification loss for sample \({\varvec{x}}_i\). The summation of this loss over all training samples \(\sum _{i=1}^{N}\mathcal {L}_{C}({\varvec{y}}_i,\hat{{\varvec{y}}}_i)\) could be used for optimization.
2.3 Optimization with Continuation
With the ranking preserving loss in (2) and the semantic classification loss in (3), we derive the overall objective for hash learning as
where \(\lambda _R\), \(\lambda _C\) and \(\lambda _p\) are hyper-parameters to balance the effects of the three terms. The third term is the regularizer term over parameters of the mapping \({\varvec{g}}\). This objective is non-differentiable due to the binary constraint of \({\varvec{b}}_i\in \{-1,1\}\) in (2), which makes the standard back-propagation method infeasible to train the deep model. With the activation of \(\text {tanh}(\cdot )\) being within \([-1,1]\), most existing works circumvent the non-smooth problem with the error-prone relaxation to approximate \(\text {sgn}\) function with \(\text {tanh}\) function. In contrast, we leverage the continuation method [7] to gradually smoothing the objective with parameterized hyperbolic tangent functions with enlarging scale parameter \(\alpha \). The \(\text {sgn}\) function can be regarded as the parameterized \(\text {tanh}\) function with infinity scale parameter
Thus, we train the network with the initial value of scale parameter \(\alpha _0\) as 1 and increase it according to the predefined sequence. For each scale parameter \(\alpha _i\), after the network converges, we use the converged network parameters to initialize the training over next scale parameter \(\alpha _{i+1}\).
3 Experiments and Results
Database: Our database builds on the NIH Chest X-ray database [15], which is currently the largest public chest X-ray database. The NIH Chest X-ray database comprises of 112,120 frontal-view X-ray images from 30,805 unique patients. Each image is with multiple labels, attached with one or more of fourteen common thoracic pathologies mined from the associated radiological reports. To build our database, we selected 13,000 images of 13 most frequent pathologies, which are Atelectasis, Consolidation, Infiltration, Pneumothorax, Edema, Emphysema, Fibrosis, Effusion, Pneumonia, Pleural_thickening, Cardiomegaly, Nodule and Mass. We constitute the training (80%) and testing (20%) sets with both patient and pathology-level non-overlapping splits to avoid positive bias.
Evaluation Settings: We compare our method with shallow learning based method: ITQ [3] and SSH [4], and deep learning based method: DH [5], SDH [5] and DSH [6]. We report their results by running the source codes provided by their respective authors to train the models by ourselves. We directly use the raw pixels as input for the convolutional neural network and 1024-D GIST feature otherwise.
In our implementation, we utilize the AlexNet network structure [12] and implement it in the Caffe [16] framework. We train the network from scratch by setting the batch size as 256, momentum as 0.9, and weight decay as 0.005. The learning rate is set to an initial value of \(10^{-4}\) with 40% decrease every 10,000 iterations. We set the length of the ranking list M as 3 to include the samples those share all, at least one and none of the labels with the query sample. For parameter tuning, we evenly split the training set into ten parts to cross validate the parameters. We set \(\rho \) as 5, \(\alpha \) as a sequence of 10 values from 1 to infinity, \(\lambda _R\) as \(10^{-1}\), \(\lambda _C\) as 1 and \(\lambda _p\) as \(10^{-4}\).
We evaluate the retrieval performance of generated binary codes with three main metrics: Normalized Discounted Cumulative Gain (NDCG) [17], Average Cumulative Gain (ACG) [17] and weighted mean Average Precision (\(\text {mAP}_\text {w}\)). NDCG for the truncated ranking list with p results is computed as \(NDCG@p=\frac{1}{Z}\sum _{i=1}^{p}\frac{2^{r_i}-1}{\log {\left( 1+i\right) }}\) where Z is a constant related to p to ensure the NDCG score for the correct order as 1. ACG is computed by \(ACG@p=\frac{1}{p}\sum _{i=1}^{p}r_i\). And \(\text {mAP}_\text {w}\) is computed by \(mAP_w=\frac{1}{Q}\sum _{q=1}^{Q}\frac{\sum _{p=1}^{M}{\delta {(r_p>0)}ACG@p}}{M_{r>0}}\) with indicator function \(\delta (\cdot )\in \{0,1\}\) and \(M_{r>0}\) being the number of relevant samples. We evaluate the performance over binary codes with lengths of 16, 32, 48, and 64 bits.
Results and Analysis: Table 1 demonstrates the retrieval performance of different hashing methods in terms of NDCG@100 for different lengths of binary codes. We can observe that OSDH consistently outperforms both deep learning based hashing methods and shallow hashing methods by 5%–11%. While the deep learning based hashing methods present higher performance than the shallow ones, our OSDH further improves the results by order sensitive loss and continuation optimization. The ranking performances of all evaluated metrics are shown in Fig. 2.
Significant gaps between our OSDH and state-of-the-art methods are observed for all ranking metrics over various lengths of binary codes. The effects of ranking preservation and multi-label classification are validated. In Fig. 3, we show some retrieved results for our OSDH. Images sharing more pathologies with the query image are preferred to be ranked at top. This indicates our OSDH is able to preserve the multilevel similarity and return images with high similarity level for better assessment assistance.
To study the effects of different terms in the objective, we perform ablative testing by setting \(\lambda _R\) as 0 (OSDH-R) or \(\lambda _C\) as 0 (OSDH-C). The performance results are listed in Table 2 for 32-bit binary codes. From the table, we can find that the multi-label classification term contributes more to the performance improvement compared against the ranking list preservation. Note that the performances of both OSDH-R and OSDH-C are higher than the performances of state-of-the-art hashing methods as reported in Table 1. Combining these two loss terms, the performance is higher than individual baselines. This implies the label information is not fully exploit by the triplet based ranking loss and the ranking list information is important to capture the multilevel similarity.
4 Conclusion
In this paper, we have proposed a learning-based hashing method for scalable multimorbidity medical image retrieval for better assessment assistance. By formulating the retrieval problem as a multi-label hash learning problem, we develop an order sensitive deep hashing method to capture the multilevel semantic similarity by both ranking list preservation and multi-label classification. We propose to optimize the learning problem with continuation to reduce the quantization loss. We conduct extensive experiments to validate the superiority of the proposed OSDH in comparison with several state-of-the-art hashing methods.
References
Zhang, X., Liu, W., Dundar, M., Badve, S., Zhang, S.: Towards large-scale histopathological image analysis: Hashing-based image retrieval. TMI 34(2), 496–506 (2015)
Weiss, Y., Torralba, A., Fergus, R.: Spectral hashing. In: NIPS, pp. 1753–1760 (2008)
Gong, Y., Lazebnik, S., Gordo, A., Perronnin, F.: Iterative quantization: a procrustean approach to learning binary codes for large-scale image retrieval. TPAMI 35(12), 2916–2929 (2013)
Wang, J., Kumar, S., Chang, S.: Semi-supervised hashing for large-scale search. TPAMI 34(12), 2393–2406 (2012)
Liong, V.E., Lu, J., Wang, G., Moulin, P., Zhou, J.: Deep hashing for compact binary codes learning. In: CVPR, pp. 2475–2483 (2015)
Liu, H., Wang, R., Shan, S., Chen, X.: Deep supervised hashing for fast image retrieval. In: CVPR, pp. 2064–2072 (2016)
Cao, Z., Long, M., Wang, J., Yu, P.S.: Hashnet: deep learning to hash by continuation. In: ICCV, pp. 5608–5617 (2017)
Wang, J., Zhang, T., Song, J., Sebe, N., Shen, H.T.: A survey on learning to hash. CoRR abs/1606.00185 (2016)
Conjeti, S., Paschali, M., Katouzian, A., Navab, N.: Deep multiple instance hashing for scalable medical image retrieval. In: MICCAI, pp. 550–558 (2017)
Sze-To, A., Tizhoosh, H.R., Wong, A.K.C.: Binary codes for tagging x-ray images via deep de-noising autoencoders. In: IJCNN, pp. 2864–2871 (2016)
Conjeti, S., Roy, A.G., Katouzian, A., Navab, N.: Hashing with residual networks for image retrieval. In: MICCAI, pp. 541–549 (2017)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: NIPS, pp. 1106–1114 (2012)
Li, Q., Sun, Z., He, R., Tan, T.: Deep supervised discrete hashing. In: NIPS, pp. 2479–2488 (2017)
Yang, H., Lin, K., Chen, C.: Supervised learning of semantics-preserving hash via deep convolutional neural networks. TPAMI 40(2), 437–451 (2018)
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., Summers, R.M.: Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: CVPR, pp. 3462–3471 (2017)
Jia, Y., et al.: Caffe: convolutional architecture for fast feature embedding. In: ACM MM, pp. 675–678 (2014)
Järvelin, K., Kekäläinen, J.: IR evaluation methods for retrieving highly relevant documents. In: SIGIR, pp. 41–48 (2000)
Acknowledgment
This work was supported in part by the National Key Research and Development Program of China under Grant 2017YFA0700802, the National Natural Science Foundation of China under Grants 61672306, U1713214, 61572271, and 61527808, the National 1000 Young Talents Plan Program, the National Postdoctoral Program for Innovative Talents under Grant BX201700137, China Postdoctoral Science Foundation under Grant 2018M630159, Tsinghua University Initiative Scientific Research Program.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, Z., Cai, R., Lu, J., Feng, J., Zhou, J. (2018). Order-Sensitive Deep Hashing for Multimorbidity Medical Image Retrieval. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_70
Download citation
DOI: https://doi.org/10.1007/978-3-030-00928-1_70
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00927-4
Online ISBN: 978-3-030-00928-1
eBook Packages: Computer ScienceComputer Science (R0)