Abstract
This paper presents a method to generate a volumetric phantom with internal anatomical structures from the patient’s skin surface geometry, and studies the potential impact of this technology on planning medical scans and procedures such as patient positioning. Existing scan planning for imaging is either done by visual inspection of the patient or based on an ionizing scan obtained prior to the full scan. These methods are either limited in accuracy or result in additional radiation dose to the patient. Our approach generates a “CT”-like phantom, with lungs and bone structures, from the patient’s skin surface. The skin surface can be estimated from a 2.5D depth sensor and thus, the proposed method offers a novel solution to reduce the radiation dose. We present quantitative experiments on a dataset of 2045 whole body CT scans and report measurements relevant to the potential clinical use of such phantoms. (This feature is based on research, and is not commercially available. Due to regulatory reasons its future availability cannot be guaranteed.)
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Medical imaging technologies such as Computed Tomography (CT) plays a pivotal role in clinical diagnosis and therapy planning. However, acquisition of CT data exposes patients to potentially harmful ionizing radiation. Several planning methodologies to reduce the radiation dose have been developed [7, 13]. However, existing CT scan planning is often performed based on coarse patient measurement estimates from visual inspection by the technician or using scouting scans (topograms). For certain other imaging methods such as emission based tomography (PET/SPECT), a CT scan is obtained prior to the procedure, to be used for attenuation correction [14]. Both these methods expose patients to additional radiation. In this paper, we present an approach to generate a volumetric phantom with density estimates of lungs and bone structures, from the patient’s body surface mesh.
With the recent developments in human body shape modeling and simulation, accurate and detailed body models are achievable for a wide range of applications in multimedia, safety, as well as diagnostic and therapeutic healthcare domains [10, 11]. However, existing statistical body shape modeling approaches mainly focus on the skin surface, while the healthcare domain pays more attention to the internal anatomical structures such as organs [5]. Several computational phantoms with internal anatomy have been developed over the years, particularly for the purpose of radiation dosimetry analysis [8]. Attempts have also been made to generate personalized phantoms based on patient’s physical attributes, such as body size (height, width), weight, BMI and/or gender [2, 14], which reportedly offer benefits over universal phantoms. However, these attribute measurements are often approximate, which limits the degree of personalization, thus, in turn limiting the potential clinical impact.
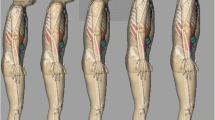
We present a learning-based framework to generate a volumetric phantom from a detailed mesh representation of the patient’s body surface; such body surface representation can be obtained using range sensors [10]. The generated phantom is a 3D volumetric image where the voxel intensity provides an estimate of the physical density based on the statistical distribution over a large dataset of patient scans. Figure 1 illustrates “ground truth” phantoms of different patients which are computed from their whole body CT scans. In this study, we focus on a phantom with lungs and bone structures, which allows evaluating our framework on its ability to generate finer details (on bones structures), while simultaneously attempting to capture the correlation between the body geometry and size/shape of lungs. During the training phase, we utilize the whole body CT scans to obtain the volumetric masks for skin, lungs, and bones, and then train a conditional deep generative network [3, 4] to learn a mapping from skin mask to a phantom with lungs and bones. Training is performed using more than 1500 whole body CT scans. Quantitative evaluations are conducted on 133 unseen patients by comparing the generated and ground truth phantoms. We also report several clinically relevant quantitative measures on phantoms which clearly demonstrates the benefits of generating phantoms from the skin surface.
2 Methods
Given the whole body CT scan of a given subject in the training dataset, we obtain the skin surface mask \(m_s\), lungs mask \(m_l\) and bone mask \(m_b\) using existing CT segmentation algorithms [1], followed by a visual validation. We define the phantom volume p as a weighted combination of these binary masks,
where \(\alpha \), \(\beta \), \(\delta \) are weights for phantom synthesis. For intensity value of the phantom voxels to be comparable to the radiodensity of the respective regions, we set these weights to be proportional to the average Hounsfield units (HU) in those regions relative to the HU of air (=\(-1000\)). In our experiments, we set \(\alpha = 1000\), \(\beta = -800\), \(\delta = 500\). Figure 1 shows phantoms for different patients.
To model the compositional nature of the phantom, we propose to use a deep network architecture that first estimates the masks for key anatomical regions (in our case, lungs, and bones) from the skin surface and then combines them into the phantom using Eq. 1. While predicting the separate masks for different regions offers the advantage of computing the losses independently and back-propagating them, it suffers from the risk that the masks may not be correlated with each other and result in physically implausible phantoms (e.g. with ribs of spine penetrating the lungs). Thus, the network architecture and training procedure must be appropriately designed to ensure that the generated phantoms are predicted with sufficiently high accuracy while ensuring physical consistency.
To achieve physical consistency, we propose to use Generative Adversarial Networks (GANs) [3]. More specifically, we employ conditional adversarial networks (cGANs) [4] which allows enforcing physical consistency without sacrificing the input output correlation (in our case, the correlation between the skin surface volume and predicted phantom). Figure 2 shows the overview of the proposed framework. In the following sections, we first introduce cGANs and then describe the details on how to adapt them to the phantom generation task.
2.1 Conditional GAN
The cGAN learns a mapping \(G:\{x,z\} \rightarrow y\) from an observed image x with additional random noise z to a synthesized image y, where x is referred as a ‘real’ sample or the condition from the original dataset, y is referred as a ‘fake’ sample generated by the trained generator G, and z is the random noise to ensure the image variability. The adversarial procedure trains a generator to produce outputs that can hardly be distinguished as a ‘fake’ by the co-trained discriminator D. Unlike GAN, the discriminator in cGAN utilizes both input x and output y of the generator to determine the ‘fake’ label based on the joint distribution. The objective function of cGAN is formulated as:
where G is the generator and D is the discriminator. The optimal \(G^*\) minimizes this objective against an adversarial D that maximizes it, and it can be solved via a min-max procedure.
2.2 Phantom Generation
The phantom generation is done at two scales. At the coarser scale, the task is formulated as a segmentation task, where network generates a body segmentation into two components - lungs mask and bone mask. At the finer scale within each mask, the network generates as many details as possible. Combining the losses of these two scales, the overall loss function of phantom generation is then defined as the combination of the cGAN loss and the segmentation loss:
where G is the generator from surface mask to the phantom, and D is the corresponding discriminator to determine whether a pair of skin and phantom are from the ground truth or the synthesis of generated masks. The cGAN loss is adapted as:
where z is the random noise, \(m_s\) is skin mask, \(p^{gt}\) is the ground truth phantom and \(p^G\) is the phantom synthesized from the masks \(m_l^G\), \(m_b^G\) generated by G using Eq. 1. The segmentation loss that quantifies the similarity between the generated and ground truth segmentation is formulated as:
where \(m^{gt} = {m_l^{gt}, m_b^{gt}}\) is the ground truth mask, \(m^G = {m_l^G, m_b^G}\) is the generated mask from G, and H is the cross entropy function.
This objective function is optimized via the cGAN adversarial procedure to obtain an optimal generator \(G^*\),
2.3 Architecture
We adapt our generator and discriminator network architectures from the Image-to-Image translation [4]. Both networks use modules consisting of Convolution-InstanceNorm-ReLU. The generator is a “U-Net” [9] with a stride of 2. The size of the embedding layer is \(1 \times 1 \times 1\). Two drop-out layers serve as the random noise z. We employ a patch-based discriminator which outputs an \(N \times N \times N\) matrix. We set N to 30 with a receptive field of size \(34 \times 34 \times 34\).
3 Experiments
3.1 Experimental Setup
To evaluate our approach, we collected 2045 whole body Computed Tomography (CT) scans from patients at several different hospital sites in North America and Europe. Our dataset contains adults with age between 20 and 87, of which 45% are female. The neck to abdomen length varies from \(1243 \pm 135\) mm. We randomly select 133 patients for testing, and use the rest for training and validation. We present a thorough analysis on phantom prediction from ground truth skin surface masks, which serves as an upper bound to what may be achievable with estimated skin surfaces using range sensors [10].
Given the skin masks and phantoms, we normalize all images to a single scale using the neck and pubic symphysis body markers (since these can be estimated from the body surface data with high accuracy \({<}2\) cm) and scaled to 128.
We compare the proposed method (referred as skin2masks+GAN) with 2 baseline approaches: (i) Use voxelwise \(L_1\)-loss, to regress the phantom from skin mask (referred as skin2phantom); (ii) Use binary cross entropy loss to generate the lungs and bone masks from the skin mask, and then obtain the phantom using Eq. 1 (referred as skin2masks). For all the experiments, we employ the same “U-Net” architecture and train using Adam [6] with mini-batch SGD. Learning rate was set to \(10^{-5}\).
3.2 Phantom Generation from Skin Surface
Quantitative Analysis. For a quantitative comparison between the proposed methods and baselines, we report the mean MS-SSIM [12] and mean \(L_1\)-error in Table 1. The MS-SSIM score, which measures the visual similarity with the ground truth phantom, is much higher for skin2masks+GAN (0.9866) compared to 0.9516 for skin2phantom and 0.9533 for skin2masks. In addition, the proposed method gains the lowest \(L_1\)-loss among the three strategies as well. We attribute the improvement in performance to the conditional adversarial training. Our understanding here is that although the two masks are spatially non-overlapping and linearly composed in the phantom, there are contextually correlated. This not only makes it possible to predict organs from surfaces but also indicates the necessity of co-optimization among parts to achieve global consistency.
Qualitative Analysis. Figure 4 show images of phantoms, predicted using the 3 methods, for several different patients. Observe that the generated phantoms from the proposed pipeline (in column 3) look visually plausible with excellent details in lungs and bone structures, indicating that the trained model reasonably maintains the underlined structures. In addition, the predicted lungs and bones mostly display adaptive variation in sizes with the ground truth. We can also see that visually neither of the two baseline methods engenders comparable quality of phantoms with the proposed method (column 3). The skin2phantom produce less details especially over the bone regions. A closer, more detailed look also reveals several issues. For patient in row 1, spinal curvature is not predicted from his skin surface, which is expected; in general, the predicted spinal columns appear more straight than the real cases. Also, the predictions for relatively larger patients (row 3–4) are not as detailed, especially in the hip region. For patient in row 4, notice that the shape of the right lung in underestimated; we guess that the thicker fat layer increases the difficulty of prediction, thereby, suggesting potential limits of the approach.
3.3 Studying Clinical Relevance of the Synthesized Phantoms
Density Estimation. To study the accuracy of the predicted density estimates, we computed the maximum deviation between the mean slice density profiles (mean intensity for every slice along SI axis) of predicted and ground truth phantoms. Such profile for a patient in testing set is shown in Fig. 3(a). The maximum deviation over testing set has a mean error of \(35.73 \pm 19.01\), which is promising. However, the histogram of the maximum deviations (see Fig. 3(b)) suggests that for 3–4% cases, error may be too high (above 80).
Lungs Estimation. Over the testing data, the distance between lung top and liver top varies between 123 and 195 mm. The lung volume varies significantly with ratio between the smallest to the largest lungs at about 3.2. We obtain the left and right lung masks from the generated phantoms and measure the volume. For volume estimation, the mean percentage error is \(21.37 \pm 13.24\) and \(18.75 \pm 14.29\) for left and right lung respectively. In comparison, the error reported by attribute based phantom [14] is \(28~\pm ~8\) and \(30~\pm ~15\). We attribute this improvement to using a detailed patient surface.
For potential use in patient positioning for lung scans, we measure the error in estimating the position of the slice that marks the lung bottom. Our method achieves a remarkably low mean error of \(17.16 \pm 12.81\) mm (with max: 51, 90%: 33) and \(15.55 \pm 11.21\) mm (with max: 30, 90%: 54) for left and right lung respectively. Although the error is large for \(5\%\) of patients, the overall low mean and 90-percentile errors clearly demonstrate the potential for clinical use.
Evaluation from Estimated Skin Surface. We use [10] to estimate complete 3D skin surface mesh and provide results for the cases for which we have both depth and full body CT data (15 cases). The mean surface distance between estimated and CT skin surface is \(13.64 \pm 4.21\) mm. The MS-SSIM of the predicted phantom CT is 0.96 and average \(L_1\) loss is 17.28. The mean percentage error for lung volume estimation increases from 21% to 27% with estimated surface instead of CT surface, which is still better than using patient meta-data [14].
4 Conclusion
In this paper, we present a method to generate a volumetric phantom from the patient’s skin surface, and report various quantitative measurements that are achievable with deep learning based methods. While the generated patient specific phantom is still likely to be limited in its ability to predict the internal anatomy, but it may still be clinically more reliable for scan planning compared to technician’s visual estimates and have the potential to be used for attenuation correction in emission tomography.
References
Birkbeck, N., et al.: Lung segmentation from CT with severe pathologies using anatomical constraints. In: MICCAI (2014)
Ding, A., Mille, M.M., Liu, T., Caracappa, P.F., Xu, X.G.: Extension of RPI-adult male and female computational phantoms to obese patients and a Monte Carlo study of the effect on CT imaging dose. Phys. Med. Biol. 57(9), 2441–2459 (2012)
Goodfellow, I., et al.: Generative adversarial nets. In: NIPS (2014)
Isola, P., Zhu, J.Y., Zhou, T., Efros, A.A.: Image-to-image translation with conditional adversarial networks. In: CVPR (2017)
Khankook, A.E.: A feasibility study on the use of phantoms with statistical lung masses for determining the uncertainty in the dose absorbed by the lung from broad beams of incident photons and neutrons. J. Radiat. Res. 58(3), 313–328 (2017)
Kingma, D., Ba, J.: Adam: a method for stochastic optimization. In: ICLR (2015)
McCollough, C.H., Primak, A.N., Braun, N., Kofler, J., Yu, L., Christner, J.: Strategies for reducing radiation dose in CT. Radiol. Clin. 47, 27–40 (2009)
Na, Y.H.: Deformable adult human phantoms for radiation protection dosimetry: anthropometric data representing size distributions of adult worker populations and software algorithms. Phys. Med. Biol. 55, 3789–3811 (2010)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Singh, V., et al.: DARWIN: deformable patient avatar representation with deep image network. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 497–504. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_56
Tsoli, A., Mahmood, N., Black, M.J.: Breathing life into shape: capturing, modeling and animating 3D human breathing. ACM Trans. Graph. 33(4), 52 (2014)
Wang, Z., Simoncelli, E.P., Bovik, A.C.: Multiscale structural similarity for image quality assessment. In: IEEE Conference Record of the Thirty-Seventh Asilomar Conference on Signals, Systems and Computers, vol. 2, pp. 1398–1402 (2003)
Zacharias, C.: Pediatric CT: strategies to lower radiation dose. Am. J. Roentgenol. 200(5), 950–956 (2013)
Zhong, X., et al.: Generation of personalized computational phantoms using only patient metadata. In: IEEE Nuclear Science Symposium and Medical Imaging Conference Record (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Wu, Y. et al. (2018). Towards Generating Personalized Volumetric Phantom from Patient’s Surface Geometry. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-00928-1_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00927-4
Online ISBN: 978-3-030-00928-1
eBook Packages: Computer ScienceComputer Science (R0)