Abstract
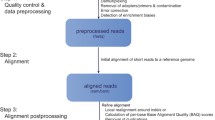
Next generation sequencing is a common and versatile tool for biological and medical research. We describe the basic steps for analyzing next generation sequencing data, including quality checking and mapping to a reference genome. We also explain the further data analysis for three common applications of next generation sequencing: variant detection, RNA-seq, and ChIP-seq.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nature Biotechnology 26:1135–1145
Medvedev P, Stanciu M, Brudno M (2009) Computational methods for discovering structural variation with next-generation sequencing. Nature Methods 6:S13-S20
Mortazavi A, Williams BA, McCue K et al (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5:621–628
Johnson DS, Mortazavi A, Myers RM et al (2007) Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science 316 (5830):1497–1502
Fu Y, Peckham HE, McLaughlin SF et al. SOLiD Sequencing and 2-Base Encoding. http://appliedbiosystems.com
Flicek P, Birney E (2009) Sense from sequence reads: methods for alignment and assembly. Nature Methods 6:S6-S12
UCSC Genome Bioinformatics. Frequently Asked Questions: Data File Formats. http://genome.ucsc.edu/FAQ/FAQformat.html
Sequence Alignment/Map (SAM) Format. http://samtools.sourceforge.net/SAM1.pdf
Korbel JO, Urban AE, Affourtit JP et al. (2007) Paired-End Mapping Reveals Extensive Structural Variation in the Human Genome. Science 318 (5849):420–426
Pepke S, Wold B, Mortazavi A (2009) Computation for ChIP-seq and RNA-seq studies. Nature Methods 6:S22-S32
Haas BJ, Zody MC (2010) Advancing RNA-seq analysis. Nature Biotechnology 28:421–423
Griffiths-Jones S, Grocock RJ, van Dongen S et al (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research 34:D140-D144. http://microrna.sanger.ac.uk
Friedländer MR, Chen W, Adamidi C et al (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nature Biotechnology 26:407–415
Database of Genomic Variants. http://projects.tcag.ca/variation
Handelsman J, Rondon MR, Brady SF et al (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & Biology 5:245–249
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Gogol-Döring, A., Chen, W. (2012). An Overview of the Analysis of Next Generation Sequencing Data. In: Wang, J., Tan, A., Tian, T. (eds) Next Generation Microarray Bioinformatics. Methods in Molecular Biology, vol 802. Humana Press. https://doi.org/10.1007/978-1-61779-400-1_16
Download citation
DOI: https://doi.org/10.1007/978-1-61779-400-1_16
Published:
Publisher Name: Humana Press
Print ISBN: 978-1-61779-399-8
Online ISBN: 978-1-61779-400-1
eBook Packages: Springer Protocols